SUMO Modification of Histone Demethylase KDM4A in Kaposi's Sarcoma-Associated Herpesvirus-Induced Primary Effusion Lymphoma
- PMID: 35914074
- PMCID: PMC9400493
- DOI: 10.1128/jvi.00755-22
SUMO Modification of Histone Demethylase KDM4A in Kaposi's Sarcoma-Associated Herpesvirus-Induced Primary Effusion Lymphoma
Abstract
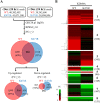
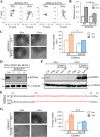
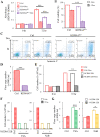
Primary effusion lymphoma (PEL) is a fatal B-cell lymphoma caused by Kaposi's sarcoma-associated herpesvirus (KSHV) infection. Inducing KSHV lytic replication that causes the death of host cells is an attractive treatment approach for PE; however, combination therapy inhibiting viral production is frequently needed to improve its outcomes. We have previously shown that the KSHV lytic protein K-bZIP can SUMOylate histone lysine demethylase 4A (KDM4A) at lysine 471 (K471) and this SUMOylation is required for virus production upon KSHV reactivation. Here, we demonstrate that SUMOylation of KDM4A orchestrates PEL cell survival, a major challenge for the success of PEL treatment; and cell movement and angiogenesis, the cell functions contributing to PEL cell extravasation and dissemination. Furthermore, integrated ChIP-seq and RNA-seq analyses identified interleukin-10 (IL-10), an immunosuppressive cytokine, as a novel downstream target of KDM4A. We demonstrate that PEL-induced angiogenesis is dependent on IL-10. More importantly, single-cell RNA sequencing (scRNA-seq) analysis demonstrated that, at the late stage of KSHV reactivation, KDM4A determines the fates of PEL cells, as evidenced by two distinct cell populations; one with less apoptotic signaling expresses high levels of viral genes and the other is exactly opposite, while KDM4A-K417R-expressing cells contain only the apoptotic population with less viral gene expression. Consistently, KDM4A knockout significantly reduced cell viability and virus production in KSHV-reactivated PEL cells. Since inhibiting PEL extravasation and eradicating KSHV-infected PEL cells without increasing viral load provide a strong rationale for treating PEL, this study indicates targeting KDM4A as a promising therapeutic option for treating PEL. IMPORTANCE PEL is an aggressive and untreatable B-cell lymphoma caused by KSHV infection. Therefore, new therapeutic approaches for PEL need to be investigated. Since simultaneous induction of KSHV reactivation and apoptosis can directly kill PEL cells, they have been applied in the treatment of this hematologic malignancy and have made progress. Epigenetic therapy with histone deacetylase (HDAC) inhibitors has been proved to treat PEL. However, the antitumor efficacies of HDAC inhibitors are modest and new approaches are needed. Following our previous report showing that the histone lysine demethylase KDM4A and its SUMOylation are required for lytic reactivation of KSHV in PEL cells, we further investigated its cellular function. Here, we found that SUMOylation of KDM4A is required for the survival, movement, and angiogenesis of lytic KSHV-infected PEL cells. Together with our previous finding showing the importance of KDM4A SUMOylation in viral production, KDM4A can be a potential therapeutic target for PEL.
Keywords: KDM4A; Kaposi's sarcoma-associated herpesvirus (KSHV); SUMOylation; histone lysine demethylase (KDM); primary effusion lymphoma (PEL).
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Lurain K, Polizzotto MN, Aleman K, Bhutani M, Wyvill KM, Goncalves PH, Ramaswami R, Marshall VA, Miley W, Steinberg SM, Little RF, Wilson W, Filie AC, Pittaluga S, Jaffe ES, Whitby D, Yarchoan R, Uldrick TS. 2019. Viral, immunologic, and clinical features of primary effusion lymphoma. Blood 133:1753–1761. 10.1182/blood-2019-01-893339. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

