Examining the epigenetic mechanisms of childhood adversity and sensitive periods: A gene set-based approach
- PMID: 35914392
- PMCID: PMC9885844
- DOI: 10.1016/j.psyneuen.2022.105854
Examining the epigenetic mechanisms of childhood adversity and sensitive periods: A gene set-based approach
Abstract
Background: Sensitive periods are developmental stages of heightened plasticity when life experiences, including exposure to childhood adversity, have the potential to exert more lasting impacts. Epigenetic mechanisms, including DNA methylation (DNAm), may provide a pathway through which adversity induces long-term biological changes. DNAm shifts may be more likely to occur during sensitive periods, especially within genes that regulate the timing of sensitive periods. Here, we investigated the possibility that childhood adversity during specific life stages is associated with DNAm changes in genes known to regulate the timing and duration of sensitive periods.
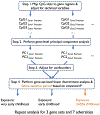
Methods: Genome-wide DNAm profiles came from the Avon Longitudinal Study of Parents and Children (n = 785). We first used principal component analysis (PCA) to summarize DNAm variation across 530 CpG sites mapped to the promoters of 58 genes previously-identified as regulating sensitive periods. Gene-level DNAm summaries were calculated for genes regulating sensitive period opening (ngenes = 15), closing (ngenes = 36), and expression (ngenes = 8). We then performed linear discriminant analysis (LDA) to test associations between seven types of parent-reported, time-varying measures of exposure to childhood adversity and DNAm principal components. To our knowledge, this is the first time LDA has been applied to analyze functionally grouped DNAm data to characterize associations between an environmental exposure and epigenetic differences.
Results: Suggestive evidence emerged for associations between sexual or physical abuse as well as financial hardship during middle childhood, and DNAm of genetic pathways regulating sensitive period opening and expression. However, no statistically significant associations were identified after multiple testing correction.
Conclusions: Our gene set-based method combining PCA and LDA complements epigenome-wide approaches. Although our results were largely null, these findings provide a proof-of-concept for studying time-varying exposures and gene- or pathway-level epigenetic modifications.
Keywords: ALSPAC; Childhood adversity; Epigenetics; Plasticity; Sensitive periods.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Sensitive Periods for the Effect of Childhood Adversity on DNA Methylation: Results From a Prospective, Longitudinal Study.Biol Psychiatry. 2019 May 15;85(10):838-849. doi: 10.1016/j.biopsych.2018.12.023. Epub 2019 Jan 21. Biol Psychiatry. 2019. PMID: 30905381 Free PMC article.
-
Adversity exposure during sensitive periods predicts accelerated epigenetic aging in children.Psychoneuroendocrinology. 2020 Mar;113:104484. doi: 10.1016/j.psyneuen.2019.104484. Epub 2019 Nov 6. Psychoneuroendocrinology. 2020. PMID: 31918390 Free PMC article.
-
Stage 2 Registered Report: Epigenetic Intergenerational Transmission: Mothers' Adverse Childhood Experiences and DNA Methylation.J Am Acad Child Adolesc Psychiatry. 2023 Oct;62(10):1110-1122. doi: 10.1016/j.jaac.2023.02.018. Epub 2023 Jun 15. J Am Acad Child Adolesc Psychiatry. 2023. PMID: 37330044 Free PMC article.
-
Epigenetics, Early Adversity and Child and Adolescent Mental Health.Psychopathology. 2018;51(2):71-75. doi: 10.1159/000486683. Epub 2018 Feb 23. Psychopathology. 2018. PMID: 29478063 Review.
-
Childhood Trauma and Epigenetics: State of the Science and Future.Curr Environ Health Rep. 2022 Dec;9(4):661-672. doi: 10.1007/s40572-022-00381-5. Epub 2022 Oct 15. Curr Environ Health Rep. 2022. PMID: 36242743 Review.
Cited by
-
Maternal adverse childhood experiences (ACEs) and offspring imprinted gene DMR methylation at birth.Epigenetics. 2024 Dec;19(1):2293412. doi: 10.1080/15592294.2023.2293412. Epub 2023 Dec 15. Epigenetics. 2024. PMID: 38100614 Free PMC article.
-
The embodiment of parental death in early life through accelerated epigenetic aging: Implications for understanding how parental death before 18 shapes age-related health risk among older adults.SSM Popul Health. 2024 Feb 29;26:101648. doi: 10.1016/j.ssmph.2024.101648. eCollection 2024 Jun. SSM Popul Health. 2024. PMID: 38596364 Free PMC article.
-
Going Beyond Childhood and Gender-Based Violence: Epigenetic Modifications and Inheritance.Womens Health Rep (New Rochelle). 2024 May 28;5(1):473-484. doi: 10.1089/whr.2024.0010. eCollection 2024. Womens Health Rep (New Rochelle). 2024. PMID: 39035135 Free PMC article. Review.
References
-
- Bach FR, Jordan MI, 2005. A Probabilistic Interpretation of Canonical Correlation Analysis 11.
-
- Battram T, Yousefi P, Crawford G, Prince C, Babei MS, Sharp G, Hatcher C, Vega-Salas MJ, Khodabakhsh S, Whitehurst O, Langdon R, Mahoney L, Elliott HR, Mancano G, Lee M, Watkins SH, Lay AC, Hemani G, Gaunt TR, Relton CL, Staley JR, Suderman M, 2021. The EWAS Catalog: a database of epigenome-wide association studies. 10.31219/osf.io/837wn - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- MC_PC_19009/MRC_/Medical Research Council/United Kingdom
- MC_UU_00011/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12013/8/MRC_/Medical Research Council/United Kingdom
- R01 HD068437/HD/NICHD NIH HHS/United States
- R01 AI121226/AI/NIAID NIH HHS/United States
- MC_UU_12013/1/MRC_/Medical Research Council/United Kingdom
- 217065/ Z/19/Z/WT_/Wellcome Trust/United Kingdom
- MRC_UU_ 00011/5/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/I025263/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBI025751/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_PC_15018/MRC_/Medical Research Council/United Kingdom
- MC_UU_00011/5/MRC_/Medical Research Council/United Kingdom
- R01 MH113930/MH/NIMH NIH HHS/United States
- MC_UU_12013/2/MRC_/Medical Research Council/United Kingdom
- G9815508/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

