Caspase Activation and Inhibition
- PMID: 35914782
- PMCID: PMC9341465
- DOI: 10.1101/cshperspect.a041020
Caspase Activation and Inhibition
Figures
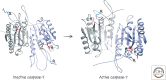
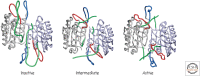

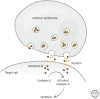
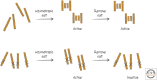
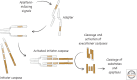
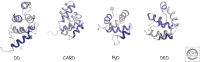
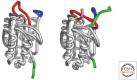

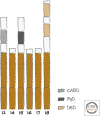
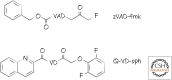

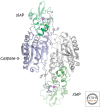
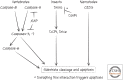
References
FIGURE CREDITS
-
- Bleakley RC. 2008. Cytotoxic T lymphocytes. In Fundamental immunology, 6th ed. (ed. Paul WE.), p. 1079. Lippincott, Williams & Wilkins, Philadelphia.
-
- Fernandes-Alnemri T, Wu J, Yu J-W, Datta P, Miller B, Jankowski W, Rosenberg S, Zhang J, Alnemri ES. 2007. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ 14: 1590–1604. 10.1038/sj.cdd.4402194 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
