PFKFB4 interacts with ICMT and activates RAS/AKT signaling-dependent cell migration in melanoma
- PMID: 35914811
- PMCID: PMC9348664
- DOI: 10.26508/lsa.202201377
PFKFB4 interacts with ICMT and activates RAS/AKT signaling-dependent cell migration in melanoma
Abstract
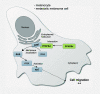
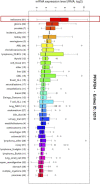
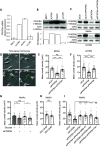
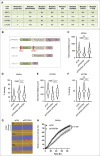
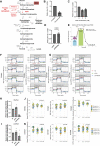
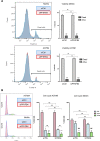
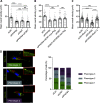
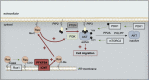
Cell migration is a complex process, tightly regulated during embryonic development and abnormally activated during cancer metastasis. RAS-dependent signaling is a major nexus controlling essential cell parameters including proliferation, survival, and migration, utilizing downstream effectors such as the PI3K/AKT signaling pathway. In melanoma, oncogenic mutations frequently enhance RAS, PI3K/AKT, or MAP kinase signaling and trigger other cancer hallmarks among which the activation of metabolism regulators. PFKFB4 is one of these critical regulators of glycolysis and of the Warburg effect. Here, however, we explore a novel function of PFKFB4 in melanoma cell migration. We find that PFKFB4 interacts with ICMT, a posttranslational modifier of RAS. PFKFB4 promotes ICMT/RAS interaction, controls RAS localization at the plasma membrane, activates AKT signaling and enhances cell migration. We thus provide evidence of a novel and glycolysis-independent function of PFKFB4 in human cancer cells. This unconventional activity links the metabolic regulator PFKFB4 to RAS-AKT signaling and impacts melanoma cell migration.
© 2022 Sittewelle et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
