Insight into the mechanism of DNA synthesis by human terminal deoxynucleotidyltransferase
- PMID: 35914812
- PMCID: PMC9348634
- DOI: 10.26508/lsa.202201428
Insight into the mechanism of DNA synthesis by human terminal deoxynucleotidyltransferase
Abstract
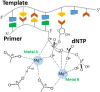
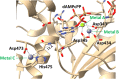
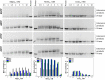
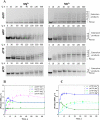
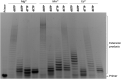
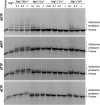
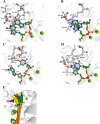
Terminal deoxynucleotidyltransferase (TdT) is a member of the DNA polymerase X family that is responsible for random addition of nucleotides to single-stranded DNA. We present investigation into the role of metal ions and specific interactions of dNTP with active-site amino acid residues in the mechanisms underlying the recognition of nucleoside triphosphates by human TdT under pre-steady-state conditions. In the elongation mode, the ratios of translocation and dissociation rate constants, as well as the catalytic rate constant were dependent on the nature of the nucleobase. Preferences of TdT in dNTP incorporation were researched by molecular dynamics simulations of complexes of TdT with a primer and dNTP or with the elongated primer. Purine nucleotides lost the "summarised" H-bonding network after the attachment of the nucleotide to the primer, whereas pyrimidine nucleotides increased the number and relative lifetime of H-bonds in the post-catalytic complex. The effect of divalent metal ions on the primer elongation revealed that Me<sup>2+</sup> cofactor can significantly change parameters of the primer elongation by strongly affecting the rate of nucleotide attachment and the polymerisation mode.
© 2022 Kuznetsova et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
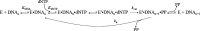



References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2: 19–25. 10.1016/j.softx.2015.06.001 - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
