The role of mitochondrial energetics in the origin and diversification of eukaryotes
- PMID: 35915152
- PMCID: PMC9575660
- DOI: 10.1038/s41559-022-01833-9
The role of mitochondrial energetics in the origin and diversification of eukaryotes
Abstract
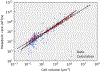
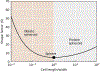
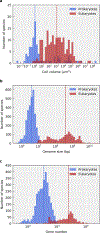
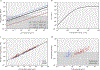
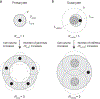
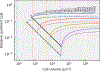
The origin of eukaryotic cell size and complexity is often thought to have required an energy excess supplied by mitochondria. Recent observations show energy demands to scale continuously with cell volume, suggesting that eukaryotes do not have higher energetic capacity. However, respiratory membrane area scales superlinearly with the cell surface area. Furthermore, the consequences of the contrasting genomic architectures between prokaryotes and eukaryotes have not been precisely quantified. Here, we investigated (1) the factors that affect the volumes at which prokaryotes become surface area-constrained, (2) the amount of energy divested to DNA due to contrasting genomic architectures and (3) the costs and benefits of respiring symbionts. Our analyses suggest that prokaryotes are not surface area-constrained at volumes of 100‒103 µm3, the genomic architecture of extant eukaryotes is only slightly advantageous at genomes sizes of 106‒107 base pairs and a larger host cell may have derived a greater advantage (lower cost) from harbouring ATP-producing symbionts. This suggests that eukaryotes first evolved without the need for mitochondria since these ranges hypothetically encompass the last eukaryotic common ancestor and its relatives. Our analyses also show that larger and faster-dividing prokaryotes would have a shortage of respiratory membrane area and divest more energy into DNA. Thus, we argue that although mitochondria may not have been required by the first eukaryotes, eukaryote diversification was ultimately dependent on mitochondria.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing interests.
Figures
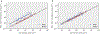


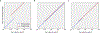

References
-
- Stanier RY, Douderoff M. & Adelberg E. The microbial world. (Prentice-Hall, 1963).
-
- Lane N. & Martin W. The energetics of genome complexity. Nature 467, 929–934 (2010). - PubMed
-
- Martin W. & Müller M. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41 (1998). - PubMed
-
- Cavalier-Smith T. Predation and eukaryote cell origins: A coevolutionary perspective. The International Journal of Biochemistry & Cell Biology 41, 307–322 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

