Cross-ancestry genome-wide meta-analysis of 61,047 cases and 947,237 controls identifies new susceptibility loci contributing to lung cancer
- PMID: 35915169
- PMCID: PMC9373844
- DOI: 10.1038/s41588-022-01115-x
Cross-ancestry genome-wide meta-analysis of 61,047 cases and 947,237 controls identifies new susceptibility loci contributing to lung cancer
Abstract
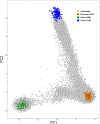
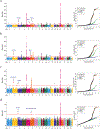
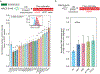
To identify new susceptibility loci to lung cancer among diverse populations, we performed cross-ancestry genome-wide association studies in European, East Asian and African populations and discovered five loci that have not been previously reported. We replicated 26 signals and identified 10 new lead associations from previously reported loci. Rare-variant associations tended to be specific to populations, but even common-variant associations influencing smoking behavior, such as those with CHRNA5 and CYP2A6, showed population specificity. Fine-mapping and expression quantitative trait locus colocalization nominated several candidate variants and susceptibility genes such as IRF4 and FUBP1. DNA damage assays of prioritized genes in lung fibroblasts indicated that a subset of these genes, including the pleiotropic gene IRF4, potentially exert effects by promoting endogenous DNA damage.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures



References
Methods-only references
Publication types
MeSH terms
Substances
Grants and funding
- HHSN268201700012C/HL/NHLBI NIH HHS/United States
- R01 CA250905/CA/NCI NIH HHS/United States
- R01 CA243483/CA/NCI NIH HHS/United States
- R03 CA256222/CA/NCI NIH HHS/United States
- U01 CA243483/CA/NCI NIH HHS/United States
- P30 CA014236/CA/NCI NIH HHS/United States
- S10 RR024574/RR/NCRR NIH HHS/United States
- T32 ES027801/ES/NIEHS NIH HHS/United States
- 001/WHO_/World Health Organization/International
- K99 ES033259/ES/NIEHS NIH HHS/United States
- R35 CA197449/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- U01 CA167462/CA/NCI NIH HHS/United States
- U19 CA203654/CA/NCI NIH HHS/United States
- P30 ES006096/ES/NIEHS NIH HHS/United States
- P30 CA076292/CA/NCI NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- DP1 AG072751/AG/NIA NIH HHS/United States
- R00 ES033259/ES/NIEHS NIH HHS/United States
- 75N92020C00001/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

