Peripheral apoE4 enhances Alzheimer's pathology and impairs cognition by compromising cerebrovascular function
- PMID: 35915180
- PMCID: PMC10009873
- DOI: 10.1038/s41593-022-01127-0
Peripheral apoE4 enhances Alzheimer's pathology and impairs cognition by compromising cerebrovascular function
Abstract
The ε4 allele of the apolipoprotein E (APOE) gene, a genetic risk factor for Alzheimer's disease, is abundantly expressed in both the brain and periphery. Here, we present evidence that peripheral apoE isoforms, separated from those in the brain by the blood-brain barrier, differentially impact Alzheimer's disease pathogenesis and cognition. To evaluate the function of peripheral apoE, we developed conditional mouse models expressing human APOE3 or APOE4 in the liver with no detectable apoE in the brain. Liver-expressed apoE4 compromised synaptic plasticity and cognition by impairing cerebrovascular functions. Plasma proteome profiling revealed apoE isoform-dependent functional pathways highlighting cell adhesion, lipoprotein metabolism and complement activation. ApoE3 plasma from young mice improved cognition and reduced vessel-associated gliosis when transfused into aged mice, whereas apoE4 compromised the beneficial effects of young plasma. A human induced pluripotent stem cell-derived endothelial cell model recapitulated the plasma apoE isoform-specific effect on endothelial integrity, further supporting a vascular-related mechanism. Upon breeding with amyloid model mice, liver-expressed apoE4 exacerbated brain amyloid pathology, whereas apoE3 reduced it. Our findings demonstrate pathogenic effects of peripheral apoE4, providing a strong rationale for targeting peripheral apoE to treat Alzheimer's disease.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures


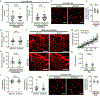





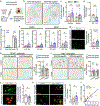


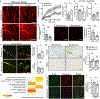



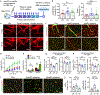


Comment in
-
Peripheral ApoE4 Leads to Cerebrovascular Dysfunction and Aβ Deposition in Alzheimer's Disease.Neurosci Bull. 2023 Aug;39(8):1330-1332. doi: 10.1007/s12264-023-01058-1. Epub 2023 Apr 24. Neurosci Bull. 2023. PMID: 37093447 Free PMC article. No abstract available.
References
-
- de la Torre JC Alzheimer disease as a vascular disorder: nosological evidence. Stroke 33, 1152–1162 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

