Target residence of Cas9-sgRNA influences DNA double-strand break repair pathway choices in CRISPR/Cas9 genome editing
- PMID: 35915475
- PMCID: PMC9341079
- DOI: 10.1186/s13059-022-02736-5
Target residence of Cas9-sgRNA influences DNA double-strand break repair pathway choices in CRISPR/Cas9 genome editing
Abstract
Background: Due to post-cleavage residence of the Cas9-sgRNA complex at its target, Cas9-induced DNA double-strand breaks (DSBs) have to be exposed to engage DSB repair pathways. Target interaction of Cas9-sgRNA determines its target binding affinity and modulates its post-cleavage target residence duration and exposure of Cas9-induced DSBs. This exposure, via different mechanisms, may initiate variable DNA damage responses, influencing DSB repair pathway choices and contributing to mutational heterogeneity in genome editing. However, this regulation of DSB repair pathway choices is poorly understood.
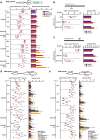
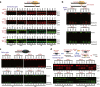
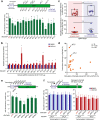
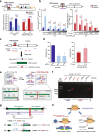
Results: In repair of Cas9-induced DSBs, repair pathway choices vary widely at different target sites and classical nonhomologous end joining (c-NHEJ) is not even engaged at some sites. In mouse embryonic stem cells, weakening the target interaction of Cas9-sgRNA promotes bias towards c-NHEJ and increases target dissociation and reduces target residence of Cas9-sgRNAs in vitro. As an important strategy for enhancing homology-directed repair, inactivation of c-NHEJ aggravates off-target activities of Cas9-sgRNA due to its weak interaction with off-target sites. By dislodging Cas9-sgRNA from its cleaved targets, DNA replication alters DSB end configurations and suppresses c-NHEJ in favor of other repair pathways, whereas transcription has little effect on c-NHEJ engagement. Dissociation of Cas9-sgRNA from its cleaved target by DNA replication may generate three-ended DSBs, resulting in palindromic fusion of sister chromatids, a potential source for CRISPR/Cas9-induced on-target chromosomal rearrangements.
Conclusions: Target residence of Cas9-sgRNA modulates DSB repair pathway choices likely through varying dissociation of Cas9-sgRNA from cleaved DNA, thus widening on-target and off-target mutational spectra in CRISPR/Cas9 genome editing.
Keywords: CRISPR-Cas9 genome editing; DNA double-strand break repair pathway choice; Editing heterogeneity; Off-target effect; Target residence.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests. Chao Lou and Hao-Dan Li are employees at Shurui Tech Ltd.
Figures



Similar articles
-
Target binding and residence: a new determinant of DNA double-strand break repair pathway choice in CRISPR/Cas9 genome editing.J Zhejiang Univ Sci B. 2021 Jan 15;22(1):73-86. doi: 10.1631/jzus.B2000282. J Zhejiang Univ Sci B. 2021. PMID: 33448189 Free PMC article. Review.
-
Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks.Int J Mol Sci. 2020 Sep 4;21(18):6461. doi: 10.3390/ijms21186461. Int J Mol Sci. 2020. PMID: 32899704 Free PMC article. Review.
-
Proximal binding of dCas9 at a DNA double strand break stimulates homology-directed repair as a local inhibitor of classical non-homologous end joining.Nucleic Acids Res. 2023 Apr 11;51(6):2740-2758. doi: 10.1093/nar/gkad116. Nucleic Acids Res. 2023. PMID: 36864759 Free PMC article.
-
Single-Strand Annealing Plays a Major Role in Double-Strand DNA Break Repair following CRISPR-Cas9 Cleavage in Leishmania.mSphere. 2019 Aug 21;4(4):e00408-19. doi: 10.1128/mSphere.00408-19. mSphere. 2019. PMID: 31434745 Free PMC article.
-
CRISPR/Cas9-Induced Double-Strand Break Repair in Arabidopsis Nonhomologous End-Joining Mutants.G3 (Bethesda). 2017 Jan 5;7(1):193-202. doi: 10.1534/g3.116.035204. G3 (Bethesda). 2017. PMID: 27866150 Free PMC article.
Cited by
-
A highly efficient method for genomic deletion across diverse lengths in thermophilic Parageobacillus thermoglucosidasius.Synth Syst Biotechnol. 2024 May 17;9(4):658-666. doi: 10.1016/j.synbio.2024.05.009. eCollection 2024 Dec. Synth Syst Biotechnol. 2024. PMID: 38817825 Free PMC article.
-
Efficient high-precision homology-directed repair-dependent genome editing by HDRobust.Nat Methods. 2023 Sep;20(9):1388-1399. doi: 10.1038/s41592-023-01949-1. Epub 2023 Jul 20. Nat Methods. 2023. PMID: 37474806 Free PMC article.
-
Characterization and regulation of cell cycle-independent noncanonical gene targeting.Nat Commun. 2024 Jun 18;15(1):5044. doi: 10.1038/s41467-024-49385-9. Nat Commun. 2024. PMID: 38890315 Free PMC article.
-
Systematic characterization of the HOXA9 downstream targets in MLL-r leukemia by noncoding CRISPR screens.Nat Commun. 2023 Nov 28;14(1):7464. doi: 10.1038/s41467-023-43264-5. Nat Commun. 2023. PMID: 38016946 Free PMC article.
-
TREX2 enables efficient genome disruption mediated by paired CRISPR-Cas9 nickases that generate 3'-overhanging ends.Mol Ther Nucleic Acids. 2023 Nov 2;34:102072. doi: 10.1016/j.omtn.2023.102072. eCollection 2023 Dec 12. Mol Ther Nucleic Acids. 2023. PMID: 38028195 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

