ZNF354C Mediated by DNMT1 Ameliorates Lung Ischemia-Reperfusion Oxidative Stress Injury by Reducing TFPI Promoter Methylation to Upregulate TFPI
- PMID: 35915612
- PMCID: PMC9338733
- DOI: 10.1155/2022/7288729
ZNF354C Mediated by DNMT1 Ameliorates Lung Ischemia-Reperfusion Oxidative Stress Injury by Reducing TFPI Promoter Methylation to Upregulate TFPI
Retraction in
-
Retracted: ZNF354C Mediated by DNMT1 Ameliorates Lung Ischemia-Reperfusion Oxidative Stress Injury by Reducing TFPI Promoter Methylation to Upregulate TFPI.Oxid Med Cell Longev. 2023 Dec 29;2023:9785630. doi: 10.1155/2023/9785630. eCollection 2023. Oxid Med Cell Longev. 2023. PMID: 38189022 Free PMC article.
Abstract
Background: Pulmonary ischemia reperfusion- (I/R-) induced dysfunction is a significant clinical problem after lung transplantation. In this study, we aim to explore the molecular mechanism of lung I/R injury (LIRI).
Methods: Bioinformatic analysis of gene involved in oxidative stress. A HUVEC oxygen glucose deprivation/reoxygenation (OGD/R) model and I/R mouse model were first established via I/R. The cellular proliferation, migration, reactive oxygen species (ROS), and parameters of lung injury were assessed via CCK-8, EdU staining, Transwell, cellular ROS kit, and H&E staining. We also confirmed related gene expressions and protein levels and the interaction between the tissue factor pathway inhibitor (TFPI) promotor and ZNF354C.
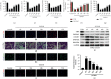
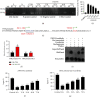
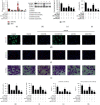
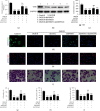
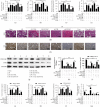
Results: Bioinformatic analysis results showed TFPI contributed to oxidative stress. OGD/R caused a reduction in cell viability and migration, hypermethylation of TFPI, increased ROS, and downregulation of ZNF354C, TFPI, and DNA methyltransferases (DNMTs) in HUVECs. Besides, ZNF354C could directly bind to the TFPI promoter, enhance proliferation and migration, and inhibit ROS in OGD/R-induced HUVECs by upregulating TFPI. More importantly, we discovered that 5-Aza could reduce TFPI methylation, upregulate TFPI, and enhance the binding of ZNF354C to the TFPI promoter in LIRI. Furthermore, DNMT1 silencing could induce proliferation and migration and prevent ROS in OGD/R-induced HUVECs by upregulating ZNF354C. Additionally, we verified that ZNF354C could alleviate LIRI by preventing DNA methylation in vivo.
Conclusions: ZNF354C overexpression induced proliferation and migration, as well as suppressed ROS in OGD/R-induced HUVECs, and alleviated LIRI in mice by inhibiting TFPI promoter methylation to upregulate TFPI. Therefore, ZNF354C and TFPI methylation might be promising molecular markers for LIRI therapy.
Copyright © 2022 Qi Shi et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Ovechkin A. V., Lominadze D., Sedoris K. C., Robinson T. W., Tyagi S. C., Roberts A. M. Lung ischemia-reperfusion injury: implications of oxidative stress and platelet-arteriolar wall interactions. Archives of Physiology and Biochemistry . 2007;113(1):1–12. doi: 10.1080/13813450601118976. - DOI - PMC - PubMed
-
- den Hengst W. A., Gielis J. F., Lin J. Y., Van Schil P. E., De Windt L. J., Moens A. L. Lung ischemia-reperfusion injury: a molecular and clinical view on a complex pathophysiological process. American journal of physiology Heart and circulatory physiology . 2010;299(5):H1283–H1299. doi: 10.1152/ajpheart.00251.2010. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

