Inhibition of c-MYC-miRNA 19 Pathway Sensitized CML K562 Cells to Etoposide via NHE1 Upregulation
- PMID: 35915613
- PMCID: PMC9338868
- DOI: 10.1155/2022/9306614
Inhibition of c-MYC-miRNA 19 Pathway Sensitized CML K562 Cells to Etoposide via NHE1 Upregulation
Abstract
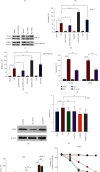
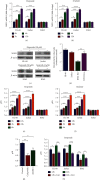
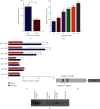
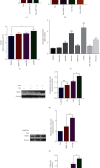
As a previously discovered target of DNA damage, Na+/H+ exchanger 1 (NHE1) plays a role in regulation of intracellular pH (pHi) through the extrusion of intracellular proton (H+) in exchange for extracellular sodium (Na+). Its abnormal expression and dysfunction have been reported in solid tumor and hematopoietic malignancies. Here, we reported that suppression of NHE1 in BCR-ABL+ hematopoietic malignancies' K562 cells treated with Etoposide was manipulated by miR-19 and c-MYC. Inhibition of miR-19 or c-MYC enhanced the expression of NHE1 and sensitized K562 cells to Etoposide in vitro. The in vivo nude mouse transplantation model was also performed to confirm the enhanced sensitivity of K562 cells to Etoposide by inhibiting the miR-19 or c-MYC pathway. TCGA analysis conferred a negative correlation between miR-19 level and leukemia patients' survival. Thus, our results provided a potential management by which the c-MYC-miRNA 19 pathway might have a crucial impact on sensitizing K562 cells to Etoposide in the therapeutic approaches.
Copyright © 2022 Shannan Cao et al.
Conflict of interest statement
We state here that none of our authors has financial or other conflicts of interest that might be constructed as influencing the results or interpretation of our study.
Figures

Similar articles
-
Induction of heme oxygenase-1 by Na+-H+ exchanger 1 protein plays a crucial role in imatinib-resistant chronic myeloid leukemia cells.J Biol Chem. 2015 May 15;290(20):12558-71. doi: 10.1074/jbc.M114.626960. Epub 2015 Mar 23. J Biol Chem. 2015. PMID: 25802333 Free PMC article. Clinical Trial.
-
Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells.Blood. 2007 May 15;109(10):4399-405. doi: 10.1182/blood-2006-09-045104. Epub 2007 Feb 6. Blood. 2007. PMID: 17284533
-
Upregulation of miR-181c inhibits chemoresistance by targeting ST8SIA4 in chronic myelocytic leukemia.Oncotarget. 2016 Sep 13;7(37):60074-60086. doi: 10.18632/oncotarget.11054. Oncotarget. 2016. PMID: 27527856 Free PMC article.
-
A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA.Oncogene. 2015 Apr 2;34(14):1768-79. doi: 10.1038/onc.2014.131. Epub 2014 May 19. Oncogene. 2015. PMID: 24837367
-
Novel agent nitidine chloride induces erythroid differentiation and apoptosis in CML cells through c-Myc-miRNAs axis.PLoS One. 2015 Feb 3;10(2):e0116880. doi: 10.1371/journal.pone.0116880. eCollection 2015. PLoS One. 2015. PMID: 25647305 Free PMC article.
References
-
- Zeller C., Hinzmann B., Seitz S., et al. SASH1: a candidate tumor suppressor gene on chromosome 6q24.3 is downregulated in breast cancer. Oncogene . 2003;22(19):2972–2983. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

