Distance decay 2.0 - A global synthesis of taxonomic and functional turnover in ecological communities
- PMID: 35915625
- PMCID: PMC9322010
- DOI: 10.1111/geb.13513
Distance decay 2.0 - A global synthesis of taxonomic and functional turnover in ecological communities
Abstract
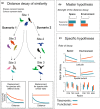
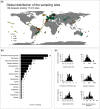
Aim: Understanding the variation in community composition and species abundances (i.e., β-diversity) is at the heart of community ecology. A common approach to examine β-diversity is to evaluate directional variation in community composition by measuring the decay in the similarity among pairs of communities along spatial or environmental distance. We provide the first global synthesis of taxonomic and functional distance decay along spatial and environmental distance by analysing 148 datasets comprising different types of organisms and environments.
Location: Global.
Time period: 1990 to present.
Major taxa studied: From diatoms to mammals.
Method: We measured the strength of the decay using ranked Mantel tests (Mantel r) and the rate of distance decay as the slope of an exponential fit using generalized linear models. We used null models to test whether functional similarity decays faster or slower than expected given the taxonomic decay along the spatial and environmental distance. We also unveiled the factors driving the rate of decay across the datasets, including latitude, spatial extent, realm and organismal features.
Results: Taxonomic distance decay was stronger than functional distance decay along both spatial and environmental distance. Functional distance decay was random given the taxonomic distance decay. The rate of taxonomic and functional spatial distance decay was fastest in the datasets from mid-latitudes. Overall, datasets covering larger spatial extents showed a lower rate of decay along spatial distance but a higher rate of decay along environmental distance. Marine ecosystems had the slowest rate of decay along environmental distances.
Main conclusions: In general, taxonomic distance decay is a useful tool for biogeographical research because it reflects dispersal-related factors in addition to species responses to climatic and environmental variables. Moreover, functional distance decay might be a cost-effective option for investigating community changes in heterogeneous environments.
Keywords: biogeography; environmental gradient; spatial distance; trait; β‐diversity.
© 2022 The Authors. Global Ecology and Biogeography published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Assessing the equilibrium between assemblage composition and climate: A directional distance-decay approach.J Anim Ecol. 2021 Aug;90(8):1906-1918. doi: 10.1111/1365-2656.13509. Epub 2021 May 20. J Anim Ecol. 2021. PMID: 33909913
-
To what extent does Tobler's 1st law of geography apply to macroecology? A case study using American palms (Arecaceae).BMC Ecol. 2008 May 22;8:11. doi: 10.1186/1472-6785-8-11. BMC Ecol. 2008. PMID: 18498661 Free PMC article.
-
Decay of similarity across tropical forest communities: integrating spatial distance with soil nutrients.Ecology. 2022 Feb;103(2):e03599. doi: 10.1002/ecy.3599. Epub 2021 Dec 28. Ecology. 2022. PMID: 34816429
-
The latitudinal gradient in plant community assembly processes: A meta-analysis.Ecol Lett. 2022 Jul;25(7):1711-1724. doi: 10.1111/ele.14019. Epub 2022 May 26. Ecol Lett. 2022. PMID: 35616424 Review.
-
Measuring biodiversity to explain community assembly: a unified approach.Biol Rev Camb Philos Soc. 2011 Nov;86(4):792-812. doi: 10.1111/j.1469-185X.2010.00171.x. Epub 2010 Dec 14. Biol Rev Camb Philos Soc. 2011. PMID: 21155964 Review.
Cited by
-
Tree phytochemical diversity and herbivory are higher in the tropics.Nat Ecol Evol. 2024 Aug;8(8):1426-1436. doi: 10.1038/s41559-024-02444-2. Epub 2024 Jun 27. Nat Ecol Evol. 2024. PMID: 38937611
-
Macroalgal microbiome biogeography is shaped by environmental drivers rather than geographical distance.Ann Bot. 2024 Mar 8;133(1):169-182. doi: 10.1093/aob/mcad151. Ann Bot. 2024. PMID: 37804485 Free PMC article.
-
Detection of functional diversity gradients and their geoclimatic filters is sensitive to data types (occurrence vs. abundance) and spatial scales (sites vs. regions).Plant Divers. 2024 Jul 4;46(6):732-743. doi: 10.1016/j.pld.2024.06.004. eCollection 2024 Nov. Plant Divers. 2024. PMID: 39811812 Free PMC article.
-
Biotic interactions and environmental filtering both determine earthworm alpha and beta diversity in tropical rainforests.Oecologia. 2025 Sep 1;207(9):151. doi: 10.1007/s00442-025-05788-z. Oecologia. 2025. PMID: 40890382
-
Spatiotemporal dynamics reveal high turnover and contrasting assembly processes in fungal communities across contiguous habitats of tropical forests.Environ Microbiome. 2025 Feb 15;20(1):23. doi: 10.1186/s40793-025-00683-9. Environ Microbiome. 2025. PMID: 39955594 Free PMC article.
References
-
- Anderson, M. J. , Crist, T. O. , Chase, J. M. , Vellend, M. , Inouye, B. D. , Freestone, A. L. , Sanders, N. J. , Cornell, H. V. , Comita, L. S. , Davies, K. F. , Harrison, S. P. , Kraft, N. J. B. , Stegen, J. C. , & Swenson, N. G. (2011). Navigating the multiple meanings of β diversity: A roadmap for the practicing ecologist. Ecology Letters, 14, 19–28. 10.1111/j.1461-0248.2010.01552.x - DOI - PubMed
-
- Astorga, A. , Oksanen, J. , Luoto, M. , Soininen, J. , Virtanen, R. , & Muotka, T. (2012). Distance decay of similarity in freshwater communities: Do macro‐ and microorganisms follow the same rules? Global Ecology and Biogeography, 21, 365–375. 10.1111/j.1466-8238.2011.00681.x - DOI
-
- Bagaria, G. , Pino, J. , Rodà, F. , & Guardiola, M. (2012). Species traits weakly involved in plant responses to landscape properties in Mediterranean grasslands. Journal of Vegetation Science, 23, 432–442. 10.1111/j.1654-1103.2011.01363.x - DOI
-
- Barbaro, L. , Brockerhoff, E. G. , Giffard, B. , & van Halder, I. (2012). Edge and area effects on avian assemblages and insectivory in fragmented native forests. Landscape Ecology, 27, 1451–1463. 10.1007/s10980-012-9800-x - DOI
-
- Barbaro, L. , Rusch, A. , Muiruri, E. W. , Gravellier, B. , Thiery, D. , & Castagneyrol, B. (2017). Avian pest control in vineyards is driven by interactions between bird functional diversity and landscape heterogeneity. Journal of Applied Ecology, 54, 500–508. 10.1111/1365-2664.12740 - DOI
LinkOut - more resources
Full Text Sources
