Genetically Engineered Viral Vectors and Organic-Based Non-Viral Nanocarriers for Drug Delivery Applications
- PMID: 35916145
- PMCID: PMC11481035
- DOI: 10.1002/adhm.202201583
Genetically Engineered Viral Vectors and Organic-Based Non-Viral Nanocarriers for Drug Delivery Applications
Abstract
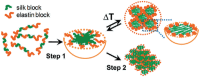
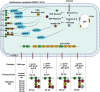
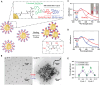
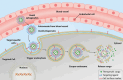
Conventional drug delivery systems are challenged by concerns related to systemic toxicity, repetitive doses, drug concentrations fluctuation, and adverse effects. Various drug delivery systems are developed to overcome these limitations. Nanomaterials are employed in a variety of biomedical applications such as therapeutics delivery, cancer therapy, and tissue engineering. Physiochemical nanoparticle assembly techniques involve the application of solvents and potentially harmful chemicals, commonly at high temperatures. Genetically engineered organisms have the potential to be used as promising candidates for greener, efficient, and more adaptable platforms for the synthesis and assembly of nanomaterials. Genetically engineered carriers are precisely designed and constructed in shape and size, enabling precise control over drug attachment sites. The high accuracy of these novel advanced materials, biocompatibility, and stimuli-responsiveness, elucidate their emerging application in controlled drug delivery. The current article represents the research progress in developing various genetically engineered carriers. Organic-based nanoparticles including cellulose, collagen, silk-like polymers, elastin-like protein, silk-elastin-like protein, and inorganic-based nanoparticles are discussed in detail. Afterward, viral-based carriers are classified, and their potential for targeted therapeutics delivery is highlighted. Finally, the challenges and prospects of these delivery systems are concluded.
Keywords: abiotic nanomaterials; genetically manipulation; nanoparticles; non-viral vectors; surface modifications.
© 2022 The Authors. Advanced Healthcare Materials published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Chamundeeswari M., Jeslin J. G., Verma M. L., Environ. Chem. Lett. 2018, 17, 849.
-
- Tabasi H., Babaei M., Abnous K., Taghdisi S. M., Saljooghi A. S., Ramezani M., Alibolandi M., J. Nanostruct. Chem. 2021, 11, 501.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

