Deep Population Genomics Reveals Systematic and Parallel Evolution at a Lipopolysaccharide Biosynthetic Locus in Xanthomonas Pathogens That Infect Rice and Sugarcane
- PMID: 35916503
- PMCID: PMC9397109
- DOI: 10.1128/aem.00550-22
Deep Population Genomics Reveals Systematic and Parallel Evolution at a Lipopolysaccharide Biosynthetic Locus in Xanthomonas Pathogens That Infect Rice and Sugarcane
Abstract
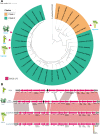
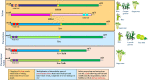
The advent of high-throughput sequencing and population genomics has enabled researchers to investigate selection pressure at hypervariable genomic loci encoding pathogen-associated molecular pattern (PAMP) molecules like lipopolysaccharide (LPS). Xanthomonas is a model and a major group of phytopathogenic bacteria that infect hosts in tissue-specific manner. Our in-depth population-based genomic investigation revealed the emergence of major lineages in two Xanthomonas pathogens that infect xylem of rice and sugarcane is associated with the acquisition and later large-scale replacement by distinct type of LPS cassettes. In the population of the rice xylem pathogen, Xanthomonas oryzae pv. oryzae (Xoo) and sugarcane pathogens Xanthomonas sacchari (Xsac) and Xanthomonas vasicola (Xvv), the BXO8 type of LPS cassette is replaced by a BXO1 type of cassette in Xoo and by Xvv type LPS cassette in Xsac and Xvv. These findings suggest a wave of parallel evolution at an LPS locus mediated by horizontal gene transfer (HGT) events during its adaptation and emergence. Aside from xylem pathogens, two closely related lineages of Xoo that infect parenchyma of rice and Leersia hexandra grass have acquired an LPS cassette from Xanthomonas pathogens that infect parenchyma of citrus, walnut, and strawberries, indicating yet another instance of parallel evolution mediated by HGT at an LPS locus. Our targeted and megapopulation-based genome dynamic studies revealed the acquisition and dominance of specific types of LPS cassettes in adaptation and success of a major group of phytopathogenic bacteria. IMPORTANCE Lipopolysaccharide (LPS) is a major microbe associated molecular pattern and hence a major immunomodulator. As a major and outer member component, it is expected that LPS is a frontline defense mechanism to deal with different host responses. Limited studies have indicated that LPS loci are also highly variable at strain and species level in plant-pathogenic bacteria, suggesting strong selection pressure from plants and associated niches. The advent of high-throughput genomics has led to the availability of a large set of genomic resources at taxonomic and population levels. This provides an exciting and important opportunity to carryout megascale targeted and population-based comparative genomic/association studies at important loci like those encoding LPS biosynthesis to understand their role in the evolution of the host, tissue specificity, and also predominant lineages. Such studies will also fill major gap in understanding host and tissue specificity in pathogenic bacteria. Our pioneering study uses the Xanthomonas group of phytopathogens that are known for their characteristic host and tissue specificity. The present deep phylogenomics of diverse Xanthomonas species and its members revealed lineage association and dominance of distinct types of LPS in accordance with their origin, host, tissue specificity, and evolutionary success.
Keywords: LPS; Xanthomonas citri; Xanthomonas oryzae; genomics; grasses; host specificity; parenchyma; pathovars; phages; phylogeny; population; tissue specificity; xylem.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Comparative genomics-based insights into Xanthomonas indica, a non-pathogenic species of healthy rice microbiome with bioprotection function.Appl Environ Microbiol. 2024 Sep 18;90(9):e0084824. doi: 10.1128/aem.00848-24. Epub 2024 Aug 19. Appl Environ Microbiol. 2024. PMID: 39158313 Free PMC article.
-
The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers.BMC Evol Biol. 2007 Dec 6;7:243. doi: 10.1186/1471-2148-7-243. BMC Evol Biol. 2007. PMID: 18053269 Free PMC article.
-
Variation suggestive of horizontal gene transfer at a lipopolysaccharide (lps) biosynthetic locus in Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen of rice.BMC Microbiol. 2004 Oct 9;4:40. doi: 10.1186/1471-2180-4-40. BMC Microbiol. 2004. PMID: 15473911 Free PMC article.
-
Xanthomonas as a Model System for Studying Pathogen Emergence and Evolution.Phytopathology. 2024 Jul;114(7):1433-1446. doi: 10.1094/PHYTO-03-24-0084-RVW. Epub 2024 Jul 5. Phytopathology. 2024. PMID: 38648116 Review.
-
Using Ecology, Physiology, and Genomics to Understand Host Specificity in Xanthomonas.Annu Rev Phytopathol. 2016 Aug 4;54:163-87. doi: 10.1146/annurev-phyto-080615-100147. Epub 2016 Jan 1. Annu Rev Phytopathol. 2016. PMID: 27296145 Review.
Cited by
-
Comparative genomics-based insights into Xanthomonas indica, a non-pathogenic species of healthy rice microbiome with bioprotection function.Appl Environ Microbiol. 2024 Sep 18;90(9):e0084824. doi: 10.1128/aem.00848-24. Epub 2024 Aug 19. Appl Environ Microbiol. 2024. PMID: 39158313 Free PMC article.
-
Strain of Xanthomonas oryzae pv. oryzae Loses Virulence through Dysregulation of Cardiolipin Synthase.Plants (Basel). 2024 Sep 14;13(18):2576. doi: 10.3390/plants13182576. Plants (Basel). 2024. PMID: 39339552 Free PMC article.
References
-
- Alexander C, Rietschel ET. 2001. Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res 7:167–202. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

