Reduction of a Heme Cofactor Initiates N-Nitroglycine Degradation by NnlA
- PMID: 35916514
- PMCID: PMC9397103
- DOI: 10.1128/aem.01023-22
Reduction of a Heme Cofactor Initiates N-Nitroglycine Degradation by NnlA
Abstract
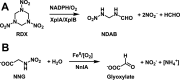
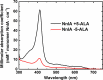
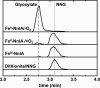
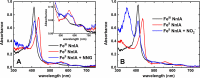
Linear nitramines are potentially carcinogenic environmental contaminants. The NnlA enzyme from Variovorax sp. strain JS1663 degrades the nitramine N-nitroglycine (NNG)-a natural product produced by some bacteria-to glyoxylate and nitrite (NO2-). Ammonium (NH4+) was predicted as the third product of this reaction. A source of nonheme FeII was shown to be required for initiation of NnlA activity. However, the role of this FeII for NnlA activity was unclear. This study reveals that NnlA contains a b-type heme cofactor. Reduction of this heme-either by a nonheme iron source or dithionite-is required to initiate NnlA activity. Therefore, FeII is not an essential substrate for holoenzyme activity. Our data show that reduced NnlA (FeII-NnlA) catalyzes at least 100 turnovers and does not require O2. Finally, NH4+ was verified as the third product, accounting for the complete nitrogen mass balance. Size exclusion chromatography showed that NnlA is a dimer in solution. Additionally, FeII-NnlA is oxidized by O2 and NO2- and stably binds carbon monoxide (CO) and nitric oxide (NO). These are characteristics shared with heme-binding PAS domains. Furthermore, a structural homology model of NnlA was generated using the PAS domain from Pseudomonas aeruginosa Aer2 as a template. The structural homology model suggested His73 is the axial ligand of the NnlA heme. Site-directed mutagenesis of His73 to alanine decreased the heme occupancy of NnlA and eliminated NNG activity, validating the homology model. We conclude that NnlA forms a homodimeric heme-binding PAS domain protein that requires reduction for initiation of the activity. IMPORTANCE Linear nitramines are potential carcinogens. These compounds result from environmental degradation of high-energy cyclic nitramines and as by-products of carbon capture technologies. Mechanistic understanding of the biodegradation of these compounds is critical to inform strategies for their remediation. Biodegradation of NNG by NnlA from Variovorax sp. strain JS 1663 requires nonheme iron, but its role is unclear. This study shows that nonheme iron is unnecessary. Instead, our study reveals that NnlA contains a heme cofactor, the reduction of which is critical for activating NNG degradation activity. These studies constrain the proposals for NnlA reaction mechanisms, thereby informing mechanistic studies of degradation of anthropogenic nitramine contaminants. In addition, these results will inform future work to design biocatalysts to degrade these nitramine contaminants.
Keywords: N-nitroglycine; PAS domain; biodegradation; enzymology; heme; nitramine; nitrogen; nitrogen metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Låg M, Lindeman B, Instanes C, Brunborg G, Schwarze PE. 2011. Health effects of amines and derivatives associated with CO2 capture. Norwegian Institute of Public Health, Norway.
-
- Chen X, Huang G, An C, Yao Y, Zhao S. 2018. Emerging N-nitrosamines and N-nitramines from amine-based post-combustion CO2 capture–a review. Chem Eng J 335:921–935. 10.1016/j.cej.2017.11.032. - DOI
-
- Rylott EL, Jackson RG, Sabbadin F, Seth-Smith HM, Edwards J, Chong CS, Strand SE, Grogan G, Bruce NC. 2011. The explosive-degrading cytochrome P450 XplA: biochemistry, structural features and prospects for bioremediation. Biochim Biophys Acta 1814:230–236. 10.1016/j.bbapap.2010.07.004. - DOI - PubMed
-
- Jenkins TF, Bartolini C, Ranney TA. 2003. Stability of CL-20, TNAZ, HMX, RDX, NG, and PETN in moist, unsaturated soil. US Army Engineer Research and Development Center, Cold Regions Research and Engineering Lab, Hanover, NH.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

