New Roles for HAMP Domains: the Tri-HAMP Region of Pseudomonas aeruginosa Aer2 Controls Receptor Signaling and Cellular Localization
- PMID: 35916529
- PMCID: PMC9487508
- DOI: 10.1128/jb.00225-22
New Roles for HAMP Domains: the Tri-HAMP Region of Pseudomonas aeruginosa Aer2 Controls Receptor Signaling and Cellular Localization
Abstract
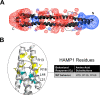
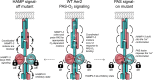
The Aer2 chemoreceptor from Pseudomonas aeruginosa is an O2 sensor involved in stress responses, virulence, and tuning the behavior of the chemotaxis (Che) system. Aer2 is the sole receptor of the Che2 system. It is soluble, but membrane associated, and forms complexes at the cell pole during stationary phase. The domain arrangement of Aer2 is unusual, with a PAS sensing domain sandwiched between five HAMP domains, followed by a C-terminal kinase-control output domain. The first three HAMP domains form a poly-HAMP chain N-terminal to the PAS sensing domain. HAMP domains are often located between signal input and output domains, where they transduce signals. Given that HAMP1 to 3 reside N-terminal to the input-output pathway, we undertook a systematic examination of their function in Aer2. We found that HAMP1 to 3 influence PAS signaling over a considerable distance, as the majority of HAMP1, 2 and 3 mutations, and deletions of helical phase stutters, led to nonresponsive signal-off or off-biased receptors. PAS signal-on lesions that mimic activated Aer2 also failed to override N-terminal HAMP signal-off replacements. This indicates that HAMP1 to 3 are critical coupling partners for PAS signaling and likely function as a cohesive unit and moveable scaffold to correctly orient and poise PAS dimers for O2-mediated signaling in Aer2. HAMP1 additionally controlled the clustering and polar localization of Aer2 in P. aeruginosa. Localization was not driven by HAMP1 charge, and HAMP1 signal-off mutants still localized. Employing HAMP as a clustering and localization determinant, as well as a facilitator of PAS signaling, are newly recognized roles for HAMP domains. IMPORTANCE P. aeruginosa is an opportunistic pathogen that interprets environmental stimuli via 26 chemoreceptors that signal through 4 distinct chemosensory systems. The second chemosensory system, Che2, contains a receptor named Aer2 that senses O2 and mediates stress responses and virulence and tunes chemotactic behavior. Aer2 is membrane associated, but soluble, and has three N-terminal HAMP domains (HAMP1 to 3) that reside outside the signal input-output pathway of Aer2. In this study, we determined that HAMP1 to 3 facilitate O2-dependent signaling from the PAS sensing domain and that HAMP1 controls the formation of Aer2-containing polar foci in P. aeruginosa. Both of these are newly recognized roles for HAMP domains that may be applicable to other non-signal-transducing HAMP domains and poly-HAMP chains.
Keywords: Aer2; HAMP domain; Pseudomonas aeruginosa; cellular localization; chemoreceptor signaling; fluorescence microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Garvis S, Munder A, Ball G, de Bentzmann S, Wiehlmann L, Ewbank JJ, Tummler B, Filloux A. 2009. Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog 5:e1000540. 10.1371/journal.ppat.1000540. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

