Construction of prediction models for growth traits of soybean cultivars based on phenotyping in diverse genotype and environment combinations
- PMID: 35916715
- PMCID: PMC9358015
- DOI: 10.1093/dnares/dsac024
Construction of prediction models for growth traits of soybean cultivars based on phenotyping in diverse genotype and environment combinations
Abstract
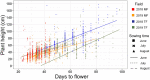
As soybean cultivars are adapted to a relatively narrow range of latitude, the effects of climate changes are estimated to be severe. To address this issue, it is important to improve our understanding of the effects of climate change by applying the simulation model including both genetic and environmental factors with their interactions (G×E). To achieve this goal, we conducted the field experiments for soybean core collections using multiple sowing times in multi-latitudinal fields. Sowing time shifts altered the flowering time (FT) and growth phenotypes, and resulted in increasing the combinations of genotypes and environments. Genome-wide association studies for the obtained phenotypes revealed the effects of field and sowing time to the significance of detected alleles, indicating the presence of G×E. By using accumulated phenotypic and environmental data in 2018 and 2019, we constructed multiple regression models for FT and growth pattern. Applicability of the constructed models was evaluated by the field experiments in 2020 including a novel field, and high correlation between the predicted and measured values was observed, suggesting the robustness of the models. The models presented here would allow us to predict the phenotype of the core collections in a given environment.
Keywords: field phenotyping; multiple regression models; multiple sowing times; soybean.
© The Author(s) 2022. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures



References
-
- Cober E.R., Morrison M.J.. 2010, Regulation of seed yield and agronomic characters by photoperiod sensitivity and growth habit genes in soybean, Theor. Appl. Genet., 120, 1005–12. - PubMed
-
- Zhang L.X., Kyei-Boahen S., Zhang J., et al.2007, Modifications of optimum adaptation zones for soybean maturity groups in the USA, Crop Manag., 6, 1–11.
-
- Deryng D., Conway D., Ramankutty N., Price J., Warren R.. 2014, Global crop yield response to extreme heat stress under multiple climate change futures, Environ. Res. Lett., 9, 034011.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

