Identification of three conserved linear B cell epitopes on the SARS-CoV-2 spike protein
- PMID: 35916768
- PMCID: PMC9487943
- DOI: 10.1080/22221751.2022.2109515
Identification of three conserved linear B cell epitopes on the SARS-CoV-2 spike protein
Abstract
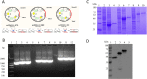
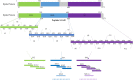
Spike (S) glycoprotein is the most significant structural protein of SARS-CoV-2 and a key target for neutralizing antibodies. In light of the on-going SARS-CoV-2 pandemic, identification and screening of epitopes of spike glycoproteins will provide vital progress in the development of sensitive and specific diagnostic tools. In the present study, NTD, RBD, and S2 genes were inserted into the pcDNA3.1(+) vector and designed with N-terminal 6× His-tag for fusion expression in HEK293F cells by transient transfection. Six monoclonal antibodies (4G, 9E, 4B, 7D, 8F, and 3D) were prepared using the expressed proteins by cell fusion technique. The characterization of mAbs was performed by indirect -ELISA, western blot, and IFA. We designed 49 overlapping synthesized peptides that cover the extracellular region of S protein in which 6 amino acid residues were offset between adjacent (S1-S49). Peptides S12, S19, and S49 were identified as the immunodominant epitope regions by the mAbs. These regions were further truncated and the peptides S12.2 286TDAVDCALDPLS297, S19.2 464FERDISTEIYQA475, and S49.4 1202ELGKYEQYIKWP1213 were identified as B- cell linear epitopes for the first time. Alanine scans showed that the D467, I468, E471, Q474, and A475 of the epitope S19.2 and K1205, Q1208, and Y1209 of the epitope S49.4 were the core sites involved in the mAbs binding. The multiple sequence alignment analysis showed that these three epitopes were highly conserved among the variants of concern (VOCs) and variants of interest (VOIs). Taken together, the findings provide a potential material for rapid diagnosis methods of COVID-19.
Keywords: B cell epitope; SARS-CoV-2; conserved epitope; monoclonal antibodies; spike protein.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




Similar articles
-
Epitope Profiling Reveals the Critical Antigenic Determinants in SARS-CoV-2 RBD-Based Antigen.Front Immunol. 2021 Sep 21;12:707977. doi: 10.3389/fimmu.2021.707977. eCollection 2021. Front Immunol. 2021. PMID: 34621266 Free PMC article.
-
Neutralizing Monoclonal Antibodies Inhibit SARS-CoV-2 Infection through Blocking Membrane Fusion.Microbiol Spectr. 2022 Apr 27;10(2):e0181421. doi: 10.1128/spectrum.01814-21. Epub 2022 Mar 16. Microbiol Spectr. 2022. PMID: 35293796 Free PMC article.
-
Screening and identification of B cell epitope of the nucleocapsid protein in SARS-CoV-2 using the monoclonal antibodies.Appl Microbiol Biotechnol. 2022 Feb;106(3):1151-1164. doi: 10.1007/s00253-022-11769-6. Epub 2022 Jan 17. Appl Microbiol Biotechnol. 2022. PMID: 35037999 Free PMC article.
-
Identification of B-Cell Epitopes for Eliciting Neutralizing Antibodies against the SARS-CoV-2 Spike Protein through Bioinformatics and Monoclonal Antibody Targeting.Int J Mol Sci. 2022 Apr 14;23(8):4341. doi: 10.3390/ijms23084341. Int J Mol Sci. 2022. PMID: 35457159 Free PMC article. Review.
-
Evolution of Anti-SARS-CoV-2 Therapeutic Antibodies.Int J Mol Sci. 2022 Aug 28;23(17):9763. doi: 10.3390/ijms23179763. Int J Mol Sci. 2022. PMID: 36077159 Free PMC article. Review.
Cited by
-
Dynamics of spike-specific neutralizing antibodies across five-year emerging SARS-CoV-2 variants of concern reveal conserved epitopes that protect against severe COVID-19.Front Immunol. 2025 Feb 18;16:1503954. doi: 10.3389/fimmu.2025.1503954. eCollection 2025. Front Immunol. 2025. PMID: 40040708 Free PMC article.
-
Display of porcine epidemic diarrhea virus spike protein B-cell linear epitope on Lactobacillus mucosae G01 S-layer surface induce a robust immunogenicity in mice.Microb Cell Fact. 2024 May 21;23(1):142. doi: 10.1186/s12934-024-02409-x. Microb Cell Fact. 2024. PMID: 38773481 Free PMC article.
References
-
- Coronavirus disease (COVID-19 Pandemic) . Available from: www.who.int/emergencies/diseases/novel-coronavirus-2019
-
- World Health Organization . Coronavirus disease (COVID-19) pandemic. [accessed 2022 Jul 12]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
