NEIL1 Recoding due to RNA Editing Impacts Lesion-Specific Recognition and Excision
- PMID: 35917336
- PMCID: PMC10231864
- DOI: 10.1021/jacs.2c03625
NEIL1 Recoding due to RNA Editing Impacts Lesion-Specific Recognition and Excision
Abstract
A-to-I RNA editing is widespread in human cells but is uncommon in the coding regions of proteins outside the nervous system. An unusual target for recoding by the adenosine deaminase ADAR1 is the pre-mRNA of the base excision DNA repair enzyme NEIL1 that results in the conversion of a lysine (K) to arginine (R) within the lesion recognition loop and alters substrate specificity. Differences in base removal by unedited (UE, K242) vs edited (Ed, R242) NEIL1 were evaluated using a series of oxidatively modified DNA bases to provide insight into the chemical and structural features of the lesion base that impact isoform-specific repair. We find that UE NEIL1 exhibits higher activity than Ed NEIL1 toward the removal of oxidized pyrimidines, such as thymine glycol, uracil glycol, 5-hydroxyuracil, and 5-hydroxymethyluracil. Gas-phase calculations indicate that the relative rates in excision track with the more stable lactim tautomer and the proton affinity of N3 of the base lesion. These trends support the contribution of tautomerization and N3 protonation in NEIL1 excision catalysis of these pyrimidine base lesions. Structurally similar but distinct substrate lesions, 5-hydroxycytosine and guanidinohydantoin, are more efficiently removed by the Ed NEIL1 isoform, consistent with the inherent differences in tautomerization, proton affinities, and lability. We also observed biphasic kinetic profiles and lack of complete base removal with specific combinations of the lesion and NEIL1 isoform, suggestive of multiple lesion binding modes. The complexity of NEIL1 isoform activity implies multiple roles for NEIL1 in safeguarding accurate repair and as an epigenetic regulator.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
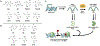

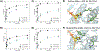


Similar articles
-
Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1.DNA Repair (Amst). 2020 Jan;85:102741. doi: 10.1016/j.dnarep.2019.102741. Epub 2019 Nov 2. DNA Repair (Amst). 2020. PMID: 31733589 Free PMC article.
-
RNA Editing of the Human DNA Glycosylase NEIL1 Alters Its Removal of 5-Hydroxyuracil Lesions in DNA.Biochemistry. 2021 May 18;60(19):1485-1497. doi: 10.1021/acs.biochem.1c00062. Epub 2021 Apr 30. Biochemistry. 2021. PMID: 33929180 Free PMC article.
-
RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1.Proc Natl Acad Sci U S A. 2010 Nov 30;107(48):20715-9. doi: 10.1073/pnas.1009231107. Epub 2010 Nov 10. Proc Natl Acad Sci U S A. 2010. PMID: 21068368 Free PMC article.
-
New paradigms in the repair of oxidative damage in human genome: mechanisms ensuring repair of mutagenic base lesions during replication and involvement of accessory proteins.Cell Mol Life Sci. 2015 May;72(9):1679-98. doi: 10.1007/s00018-014-1820-z. Epub 2015 Jan 10. Cell Mol Life Sci. 2015. PMID: 25575562 Free PMC article. Review.
-
Decoding the Role of NEIL1 Gene in DNA Repair and Lifespan: A Literature Review with Bioinformatics Analysis.Adv Biol (Weinh). 2024 Nov;8(11):e2300708. doi: 10.1002/adbi.202300708. Epub 2024 Aug 20. Adv Biol (Weinh). 2024. PMID: 39164210 Review.
Cited by
-
Zα and Zβ Localize ADAR1 to Flipons That Modulate Innate Immunity, Alternative Splicing, and Nonsynonymous RNA Editing.Int J Mol Sci. 2025 Mar 7;26(6):2422. doi: 10.3390/ijms26062422. Int J Mol Sci. 2025. PMID: 40141064 Free PMC article. Review.
-
RNA editing enzymes: structure, biological functions and applications.Cell Biosci. 2024 Mar 16;14(1):34. doi: 10.1186/s13578-024-01216-6. Cell Biosci. 2024. PMID: 38493171 Free PMC article. Review.
-
Alternative Splicing, RNA Editing, and the Current Limits of Next Generation Sequencing.Genes (Basel). 2023 Jun 30;14(7):1386. doi: 10.3390/genes14071386. Genes (Basel). 2023. PMID: 37510291 Free PMC article.
-
Research progress on the role of the NEIL family in cancer.Front Cell Dev Biol. 2025 Jul 21;13:1612329. doi: 10.3389/fcell.2025.1612329. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40761744 Free PMC article. Review.
-
Base excision repair of the N-(2-deoxy-d-erythro-pentofuranosyl)-urea lesion by the hNEIL1 glycosylase.Nucleic Acids Res. 2023 May 8;51(8):3754-3769. doi: 10.1093/nar/gkad164. Nucleic Acids Res. 2023. PMID: 37014002 Free PMC article.
References
-
- Sampath H; Batra AK; Vartanian V; Carmical JR; Prusak D; King IB; Lowell B; Earley LF; Wood TG; Marks DL; McCullough AK; R Stephen L Variable Penetrance of Metabolic Phenotypes and Development of High-Fat Diet-Induced Adiposity in NEIL1-Deficient Mice. Am. J. Physiol. Endocrinol. Metab 2011, 300 (4), E724. 10.1152/ajpendo.00387.2010. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

