Assessment of Electronic Health Record for Cancer Research and Patient Care Through a Scoping Review of Cancer Natural Language Processing
- PMID: 35917480
- PMCID: PMC9470142
- DOI: 10.1200/CCI.22.00006
Assessment of Electronic Health Record for Cancer Research and Patient Care Through a Scoping Review of Cancer Natural Language Processing
Abstract
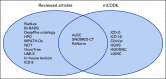
Purpose: The advancement of natural language processing (NLP) has promoted the use of detailed textual data in electronic health records (EHRs) to support cancer research and to facilitate patient care. In this review, we aim to assess EHR for cancer research and patient care by using the Minimal Common Oncology Data Elements (mCODE), which is a community-driven effort to define a minimal set of data elements for cancer research and practice. Specifically, we aim to assess the alignment of NLP-extracted data elements with mCODE and review existing NLP methodologies for extracting said data elements.
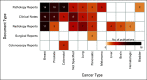
Methods: Published literature studies were searched to retrieve cancer-related NLP articles that were written in English and published between January 2010 and September 2020 from main literature databases. After the retrieval, articles with EHRs as the data source were manually identified. A charting form was developed for relevant study analysis and used to categorize data including four main topics: metadata, EHR data and targeted cancer types, NLP methodology, and oncology data elements and standards.
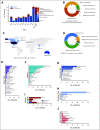
Results: A total of 123 publications were selected finally and included in our analysis. We found that cancer research and patient care require some data elements beyond mCODE as expected. Transparency and reproductivity are not sufficient in NLP methods, and inconsistency in NLP evaluation exists.
Conclusion: We conducted a comprehensive review of cancer NLP for research and patient care using EHRs data. Issues and barriers for wide adoption of cancer NLP were identified and discussed.
Conflict of interest statement
Figures



Similar articles
-
Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review.J Am Med Inform Assoc. 2019 Apr 1;26(4):364-379. doi: 10.1093/jamia/ocy173. J Am Med Inform Assoc. 2019. PMID: 30726935 Free PMC article.
-
A frame semantic overview of NLP-based information extraction for cancer-related EHR notes.J Biomed Inform. 2019 Dec;100:103301. doi: 10.1016/j.jbi.2019.103301. Epub 2019 Oct 4. J Biomed Inform. 2019. PMID: 31589927 Free PMC article. Review.
-
A scoping review of publicly available language tasks in clinical natural language processing.J Am Med Inform Assoc. 2022 Sep 12;29(10):1797-1806. doi: 10.1093/jamia/ocac127. J Am Med Inform Assoc. 2022. PMID: 35923088 Free PMC article.
-
Practical use case of natural language processing for observational clinical research data retrieval from electronic health records: AssistMED project.Pol Arch Intern Med. 2024 May 28;134(5):16704. doi: 10.20452/pamw.16704. Epub 2024 Mar 19. Pol Arch Intern Med. 2024. PMID: 38501989
-
Natural language processing with machine learning methods to analyze unstructured patient-reported outcomes derived from electronic health records: A systematic review.Artif Intell Med. 2023 Dec;146:102701. doi: 10.1016/j.artmed.2023.102701. Epub 2023 Nov 1. Artif Intell Med. 2023. PMID: 38042599 Free PMC article.
Cited by
-
DeepPhe-CR: Natural Language Processing Software Services for Cancer Registrar Case Abstraction.medRxiv [Preprint]. 2023 Oct 26:2023.05.05.23289524. doi: 10.1101/2023.05.05.23289524. medRxiv. 2023. Update in: JCO Clin Cancer Inform. 2023 Sep;7:e2300156. doi: 10.1200/CCI.23.00156. PMID: 37205575 Free PMC article. Updated. Preprint.
-
Artificial Intelligence in Cancer Research: Trends, Challenges and Future Directions.Life (Basel). 2022 Nov 28;12(12):1991. doi: 10.3390/life12121991. Life (Basel). 2022. PMID: 36556356 Free PMC article. Review.
-
Machine learning and deep learning tools for the automated capture of cancer surveillance data.J Natl Cancer Inst Monogr. 2024 Aug 1;2024(65):145-151. doi: 10.1093/jncimonographs/lgae018. J Natl Cancer Inst Monogr. 2024. PMID: 39102883 Free PMC article.
-
A comparative study of zero-shot inference with large language models and supervised modeling in breast cancer pathology classification.Res Sq [Preprint]. 2024 Feb 6:rs.3.rs-3914899. doi: 10.21203/rs.3.rs-3914899/v1. Res Sq. 2024. Update in: J Am Med Inform Assoc. 2024 Oct 1;31(10):2315-2327. doi: 10.1093/jamia/ocae146. PMID: 38405831 Free PMC article. Updated. Preprint.
-
Using natural language processing to analyze unstructured patient-reported outcomes data derived from electronic health records for cancer populations: a systematic review.Expert Rev Pharmacoecon Outcomes Res. 2024 Apr;24(4):467-475. doi: 10.1080/14737167.2024.2322664. Epub 2024 Mar 5. Expert Rev Pharmacoecon Outcomes Res. 2024. PMID: 38383308 Free PMC article.
References
-
- Tayefi M, Ngo P, Chomutare T, et al. : Challenges and opportunities beyond structured data in analysis of electronic health records. Wiley Interdiscip Rev Comput Stat 13:e1549, 2021
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

