SUMOylation inhibition overcomes proteasome inhibitor resistance in multiple myeloma
- PMID: 35917568
- PMCID: PMC9979771
- DOI: 10.1182/bloodadvances.2022007875
SUMOylation inhibition overcomes proteasome inhibitor resistance in multiple myeloma
Abstract
Proteasome inhibition is a highly effective treatment for multiple myeloma (MM). However, virtually all patients develop proteasome inhibitor resistance, which is associated with a poor prognosis. Hyperactive small ubiquitin-like modifier (SUMO) signaling is involved in both cancer pathogenesis and cancer progression. A state of increased SUMOylation has been associated with aggressive cancer biology. We found that relapsed/refractory MM is characterized by a SUMO-high state, and high expression of the SUMO E1-activating enzyme (SAE1/UBA2) is associated with poor overall survival. Consistently, continuous treatment of MM cell lines with carfilzomib (CFZ) enhanced SUMO pathway activity. Treatment of MM cell lines with the SUMO E1-activating enzyme inhibitor subasumstat (TAK-981) showed synergy with CFZ in both CFZ-sensitive and CFZ-resistant MM cell lines, irrespective of the TP53 state. Combination therapy was effective in primary MM cells and in 2 murine MM xenograft models. Mechanistically, combination treatment with subasumstat and CFZ enhanced genotoxic and proteotoxic stress, and induced apoptosis was associated with activity of the prolyl isomerase PIN1. In summary, our findings reveal activated SUMOylation as a therapeutic target in MM and point to combined SUMO/proteasome inhibition as a novel and potent strategy for the treatment of proteasome inhibitor-resistant MM.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: U.K. received speaker honorary/advisory fees from Roche, Janssen-Cilag, Takeda, BMS, Gilead, Hexal, Pfizer, AstraZeneca, PentixaPharm, Amgen, Novartis, and MSD; and clinical study support from Celgene, Takeda, BMS, Roche, AstraZeneca, Novartis, MSD, Janssen-Cilag, and Pfizer. F. Bassermann received honoraria and research funding from BMS/Celgene. J. Krönke received speaker honorary and advisory fees from BMS/Celgene and Takeda. The remaining authors declare no competing financial interests.
Figures


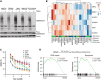

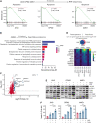
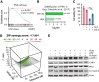
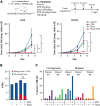
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

