Inactivated genotype 1a, 2a and 3a HCV vaccine candidates induced broadly neutralising antibodies in mice
- PMID: 35918103
- PMCID: PMC9933178
- DOI: 10.1136/gutjnl-2021-326323
Inactivated genotype 1a, 2a and 3a HCV vaccine candidates induced broadly neutralising antibodies in mice
Abstract
Objective: A prophylactic vaccine is needed to control the HCV epidemic, with genotypes 1-3 causing >80% of worldwide infections. Vaccine development is hampered by HCV heterogeneity, viral escape including protection of conserved neutralising epitopes and suboptimal efficacy of HCV cell culture systems. We developed cell culture-based inactivated genotype 1-3 HCV vaccine candidates to present natively folded envelope proteins to elicit neutralising antibodies.
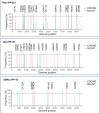
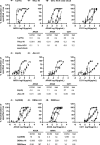
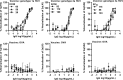
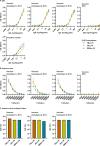
Design: High-yield genotype 1a, 2a and 3a HCV were developed by serial passage of TNcc, J6cc and DBN3acc in Huh7.5 cells and engineering of acquired mutations detected by next-generation sequencing. Neutralising epitope exposure was determined in cell-based neutralisation assays using human monoclonal antibodies AR3A and AR4A, and polyclonal antibody C211. BALB/c mice were immunised with processed and inactivated genotype 1a, 2a or 3a viruses using AddaVax, a homologue of the licenced adjuvant MF-59. Purified mouse and patient serum IgG were assayed for neutralisation capacity; mouse IgG and immune-sera were assayed for E1/E2 binding.
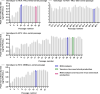
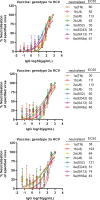
Results: Compared with the original viruses, high-yield viruses had up to ~1000 fold increased infectivity titres (peak titres: 6-7 log10 focus-forming units (FFU)/mL) and up to ~2470 fold increased exposure of conserved neutralising epitopes. Vaccine-induced IgG broadly neutralised genotype 1-6 HCV (EC50: 30-193 µg/mL; mean 71 µg/mL), compared favourably with IgG from chronically infected patients, and bound genotype 1-3 E1/E2; immune-sera endpoint titres reached up to 32 000.
Conclusion: High-yield genotype 1-3 HCV could be developed as basis for inactivated vaccine candidates inducing broadly neutralising antibodies in mice supporting further preclinical development.
Keywords: HCV; chronic viral hepatitis; hepatitis C; immune response; molecular biology.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


Similar articles
-
Inactivated whole hepatitis C virus vaccine employing a licensed adjuvant elicits cross-genotype neutralizing antibodies in mice.J Hepatol. 2022 May;76(5):1051-1061. doi: 10.1016/j.jhep.2021.12.026. Epub 2022 Jan 4. J Hepatol. 2022. PMID: 34990750
-
A Recombinant Hepatitis C Virus Genotype 1a E1/E2 Envelope Glycoprotein Vaccine Elicits Antibodies That Differentially Neutralize Closely Related 2a Strains through Interactions of the N-Terminal Hypervariable Region 1 of E2 with Scavenger Receptor B1.J Virol. 2019 Oct 29;93(22):e00810-19. doi: 10.1128/JVI.00810-19. Print 2019 Nov 15. J Virol. 2019. PMID: 31462563 Free PMC article.
-
Native Folding of a Recombinant gpE1/gpE2 Heterodimer Vaccine Antigen from a Precursor Protein Fused with Fc IgG.J Virol. 2016 Dec 16;91(1):e01552-16. doi: 10.1128/JVI.01552-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795422 Free PMC article.
-
Perspectives for a hepatitis C virus vaccine.Clin Diagn Virol. 1998 Jul 15;10(2-3):181-5. doi: 10.1016/s0928-0197(98)00028-2. Clin Diagn Virol. 1998. PMID: 9741644 Review.
-
Hepatitis C Virus Epitope Immunodominance and B Cell Repertoire Diversity.Viruses. 2021 May 25;13(6):983. doi: 10.3390/v13060983. Viruses. 2021. PMID: 34070572 Free PMC article. Review.
Cited by
-
Current Hepatitis C Vaccine Candidates Based on the Induction of Neutralizing Antibodies.Viruses. 2023 May 11;15(5):1151. doi: 10.3390/v15051151. Viruses. 2023. PMID: 37243237 Free PMC article. Review.
-
Non-cognate ligands of hepatitis C virus envelope broadly neutralizing antibodies induce virus-neutralizing sera in mice.Front Immunol. 2025 Jul 22;16:1624299. doi: 10.3389/fimmu.2025.1624299. eCollection 2025. Front Immunol. 2025. PMID: 40766312 Free PMC article.
-
Prospects for developing an Hepatitis C virus E1E2-based nanoparticle vaccine.Rev Med Virol. 2023 Sep;33(5):e2474. doi: 10.1002/rmv.2474. Epub 2023 Aug 11. Rev Med Virol. 2023. PMID: 37565536 Free PMC article. Review.
-
Bispecific antibodies against the hepatitis C virus E1E2 envelope glycoprotein.Proc Natl Acad Sci U S A. 2025 Apr 15;122(15):e2420402122. doi: 10.1073/pnas.2420402122. Epub 2025 Apr 7. Proc Natl Acad Sci U S A. 2025. PMID: 40193609
-
An inactivated SARS-CoV-2 vaccine based on a Vero cell culture-adapted high-titer virus confers cross-protection in small animals.Sci Rep. 2024 Jul 24;14(1):17039. doi: 10.1038/s41598-024-67570-0. Sci Rep. 2024. PMID: 39048693 Free PMC article.
References
-
- Hepatitis C. Available: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c [Accessed April 29, 2022].
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
