Positive strand RNA viruses differ in the constraints they place on the folding of their negative strand
- PMID: 35918125
- PMCID: PMC9479745
- DOI: 10.1261/rna.079125.122
Positive strand RNA viruses differ in the constraints they place on the folding of their negative strand
Abstract
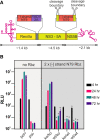
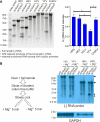
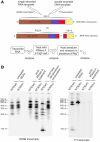
Genome replication of positive strand RNA viruses requires the production of a complementary negative strand RNA that serves as a template for synthesis of more positive strand progeny. Structural RNA elements are important for genome replication, but while they are readily observed in the positive strand, evidence of their existence in the negative strand is more limited. We hypothesized that this was due to viruses differing in their capacity to allow this latter RNA to adopt structural folds. To investigate this, ribozymes were introduced into the negative strand of different viral constructs; the expectation being that if RNA folding occurred, negative strand cleavage and suppression of replication would be seen. Indeed, this was what happened with hepatitis C virus (HCV) and feline calicivirus (FCV) constructs. However, little or no impact was observed for chikungunya virus (CHIKV), human rhinovirus (HRV), hepatitis E virus (HEV), and yellow fever virus (YFV) constructs. Reduced cleavage in the negative strand proved to be due to duplex formation with the positive strand. Interestingly, ribozyme-containing RNAs also remained intact when produced in vitro by the HCV polymerase, again due to duplex formation. Overall, our results show that there are important differences in the conformational constraints imposed on the folding of the negative strand between different positive strand RNA viruses.
Keywords: double-stranded RNA; positive strand RNA virus; replication; replication intermediate; ribozyme.
© 2022 Herod et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





Similar articles
-
Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons.FEBS J. 2005 Nov;272(22):5910-22. doi: 10.1111/j.1742-4658.2005.04986.x. FEBS J. 2005. PMID: 16279954
-
Hepatitis C Virus Replication.Cold Spring Harb Perspect Med. 2020 Mar 2;10(3):a037093. doi: 10.1101/cshperspect.a037093. Cold Spring Harb Perspect Med. 2020. PMID: 31570388 Free PMC article. Review.
-
Ribozyme gene therapy for hepatitis C virus infection.Clin Diagn Virol. 1998 Jul 15;10(2-3):163-71. doi: 10.1016/s0928-0197(98)00029-4. Clin Diagn Virol. 1998. PMID: 9741642
-
Nucleotide requirements at positions +1 to +4 for the initiation of hepatitis C virus positive-strand RNA synthesis.J Gen Virol. 2011 May;92(Pt 5):1082-1086. doi: 10.1099/vir.0.028423-0. Epub 2011 Jan 26. J Gen Virol. 2011. PMID: 21270286
-
Positive-strand RNA virus genome replication organelles: structure, assembly, control.Trends Genet. 2024 Aug;40(8):681-693. doi: 10.1016/j.tig.2024.04.003. Epub 2024 May 8. Trends Genet. 2024. PMID: 38724328 Review.
Cited by
-
Amino acid substitutions in norovirus VP1 dictate host dissemination via variations in cellular attachment.J Virol. 2023 Dec 21;97(12):e0171923. doi: 10.1128/jvi.01719-23. Epub 2023 Nov 30. J Virol. 2023. PMID: 38032199 Free PMC article.
-
The myriad roles of RNA structure in the flavivirus life cycle.RNA Biol. 2024 Jan;21(1):14-30. doi: 10.1080/15476286.2024.2357857. Epub 2024 May 26. RNA Biol. 2024. PMID: 38797925 Free PMC article. Review.
-
Structural and functional characterization of the SLA' structure at the 3' terminus of the Zika virus negative-strand intermediate.RNA. 2025 Jul 16;31(8):1139-1153. doi: 10.1261/rna.080342.124. RNA. 2025. PMID: 40341208 Free PMC article.
-
The hepatitis E virus ORF1 hypervariable region confers partial cyclophilin dependency.J Gen Virol. 2023 Nov;104(11):001919. doi: 10.1099/jgv.0.001919. J Gen Virol. 2023. PMID: 37942835 Free PMC article.
-
Thrombin cleavage of the hepatitis E virus polyprotein at multiple conserved locations is required for genome replication.PLoS Pathog. 2023 Jul 21;19(7):e1011529. doi: 10.1371/journal.ppat.1011529. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37478143 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
