Distinctive roles of translesion polymerases DinB1 and DnaE2 in diversification of the mycobacterial genome through substitution and frameshift mutagenesis
- PMID: 35918328
- PMCID: PMC9346131
- DOI: 10.1038/s41467-022-32022-8
Distinctive roles of translesion polymerases DinB1 and DnaE2 in diversification of the mycobacterial genome through substitution and frameshift mutagenesis
Abstract
Antibiotic resistance of Mycobacterium tuberculosis is exclusively a consequence of chromosomal mutations. Translesion synthesis (TLS) is a widely conserved mechanism of DNA damage tolerance and mutagenesis, executed by translesion polymerases such as DinBs. In mycobacteria, DnaE2 is the only known agent of TLS and the role of DinB polymerases is unknown. Here we demonstrate that, when overexpressed, DinB1 promotes missense mutations conferring resistance to rifampicin, with a mutational signature distinct from that of DnaE2, and abets insertion and deletion frameshift mutagenesis in homo-oligonucleotide runs. DinB1 is the primary mediator of spontaneous -1 frameshift mutations in homo-oligonucleotide runs whereas DnaE2 and DinBs are redundant in DNA damage-induced -1 frameshift mutagenesis. These results highlight DinB1 and DnaE2 as drivers of mycobacterial genome diversification with relevance to antimicrobial resistance and host adaptation.
© 2022. The Author(s).
Conflict of interest statement
M.S.G. has received consulting fees from Vedanta Biosciences, PRL NYC, and Fimbrion Therapeutics and has equity in Vedanta biosciences. All other authors declare no competing interests.
Figures
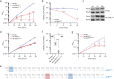

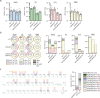


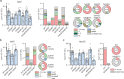

Similar articles
-
Roles for mycobacterial DinB2 in frameshift and substitution mutagenesis.Elife. 2023 May 4;12:e83094. doi: 10.7554/eLife.83094. Elife. 2023. PMID: 37141254 Free PMC article.
-
Role of the DinB homologs Rv1537 and Rv3056 in Mycobacterium tuberculosis.J Bacteriol. 2010 Apr;192(8):2220-7. doi: 10.1128/JB.01135-09. Epub 2010 Feb 5. J Bacteriol. 2010. PMID: 20139184 Free PMC article.
-
A multilayered repair system protects the mycobacterial chromosome from endogenous and antibiotic-induced oxidative damage.Proc Natl Acad Sci U S A. 2020 Aug 11;117(32):19517-19527. doi: 10.1073/pnas.2006792117. Epub 2020 Jul 29. Proc Natl Acad Sci U S A. 2020. PMID: 32727901 Free PMC article.
-
Mechanisms of mutagenesis induced by DNA lesions: multiple factors affect mutations in translesion DNA synthesis.Crit Rev Biochem Mol Biol. 2020 Jun;55(3):219-251. doi: 10.1080/10409238.2020.1768205. Epub 2020 May 24. Crit Rev Biochem Mol Biol. 2020. PMID: 32448001 Review.
-
Biological roles of translesion synthesis DNA polymerases in eubacteria.Mol Microbiol. 2010 Aug;77(3):540-8. doi: 10.1111/j.1365-2958.2010.07260.x. Epub 2010 Jun 28. Mol Microbiol. 2010. PMID: 20609084 Review.
Cited by
-
An additional proofreader contributes to DNA replication fidelity in mycobacteria.Proc Natl Acad Sci U S A. 2024 Aug 20;121(34):e2322938121. doi: 10.1073/pnas.2322938121. Epub 2024 Aug 14. Proc Natl Acad Sci U S A. 2024. PMID: 39141351 Free PMC article.
-
Newly Identified Mycobacterium africanum Lineage 10, Central Africa.Emerg Infect Dis. 2024 Oct;30(3):560-563. doi: 10.3201/eid3003.231466. Emerg Infect Dis. 2024. PMID: 38407162 Free PMC article.
-
Drug resistant tuberculosis: Implications for transmission, diagnosis, and disease management.Front Cell Infect Microbiol. 2022 Sep 23;12:943545. doi: 10.3389/fcimb.2022.943545. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36211964 Free PMC article. Review.
-
A nucleoid-associated protein is involved in the emergence of antibiotic resistance by promoting the frequent exchange of the replicative DNA polymerase in M. smegmatis.bioRxiv [Preprint]. 2024 Jan 3:2023.06.12.544663. doi: 10.1101/2023.06.12.544663. bioRxiv. 2024. Update in: mSphere. 2024 May 29;9(5):e0012224. doi: 10.1128/msphere.00122-24. PMID: 38260554 Free PMC article. Updated. Preprint.
-
The C-Terminal Acid Phosphatase Module of the RNase HI Enzyme RnhC Controls Rifampin Sensitivity and Light-Dependent Colony Pigmentation of Mycobacterium smegmatis.J Bacteriol. 2023 Apr 25;205(4):e0043122. doi: 10.1128/jb.00431-22. Epub 2023 Mar 14. J Bacteriol. 2023. PMID: 36916909 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

