Exploring synthetic lethal network for the precision treatment of clear cell renal cell carcinoma
- PMID: 35918352
- PMCID: PMC9345903
- DOI: 10.1038/s41598-022-16657-7
Exploring synthetic lethal network for the precision treatment of clear cell renal cell carcinoma
Abstract
The emerging targeted therapies have revolutionized the treatment of advanced clear cell renal cell carcinoma (ccRCC) over the past 15 years. Nevertheless, lack of personalized treatment limits the development of effective clinical guidelines and improvement of patient prognosis. In this study, large-scale genomic profiles from ccRCC cohorts were explored for integrative analysis. A credible method was developed to identify synthetic lethality (SL) pairs and a list of 72 candidate pairs was determined, which might be utilized to selectively eliminate tumors with genetic aberrations using SL partners of specific mutations. Further analysis identified BRD4 and PRKDC as novel medical targets for patients with BAP1 mutations. After mapping these target genes to the comprehensive drug datasets, two agents (BI-2536 and PI-103) were found to have considerable therapeutic potentials in the BAP1 mutant tumors. Overall, our findings provided insight into the overview of ccRCC mutation patterns and offered novel opportunities for improving individualized cancer treatment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
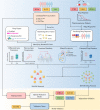

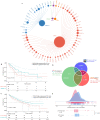
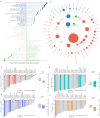
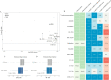
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

