Uncontrolled transposition following RNAi loss causes hypermutation and antifungal drug resistance in clinical isolates of Cryptococcus neoformans
- PMID: 35918426
- PMCID: PMC10840647
- DOI: 10.1038/s41564-022-01183-z
Uncontrolled transposition following RNAi loss causes hypermutation and antifungal drug resistance in clinical isolates of Cryptococcus neoformans
Abstract
Cryptococcus neoformans infections cause approximately 15% of AIDS-related deaths owing to a combination of limited antifungal therapies and drug resistance. A collection of clinical and environmental C. neoformans isolates were assayed for increased mutation rates via fluctuation analysis, and we identified two hypermutator C. neoformans clinical isolates with increased mutation rates when exposed to the combination of rapamycin and FK506. Sequencing of drug target genes found that Cnl1 transposon insertions conferred the majority of resistance to rapamycin and FK506 and could also independently cause resistance to 5-fluoroorotic acid and the clinically relevant antifungal 5-flucytosine. Whole-genome sequencing revealed both hypermutator genomes harbour a nonsense mutation in the RNA-interference component ZNF3 and hundreds of Cnl1 elements organized into massive subtelomeric arrays on each of the fourteen chromosomes. Quantitative trait locus mapping in 28 progeny derived from a cross between a hypermutator and wild-type identified a locus associated with hypermutation that included znf3. CRISPR editing of the znf3 nonsense mutation abolished hypermutation and restored small-interfering-RNA production. We conclude that hypermutation and drug resistance in these clinical isolates result from RNA-interference loss and accumulation of Cnl1 elements.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests.
The authors declare no competing financial or non-financial interests.
Figures





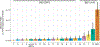

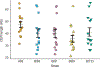

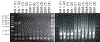





Comment in
-
Telomere transposon takeover in Cryptococcus.Nat Microbiol. 2022 Aug;7(8):1108-1109. doi: 10.1038/s41564-022-01189-7. Nat Microbiol. 2022. PMID: 35918419 No abstract available.
Similar articles
-
Distinct evolutionary trajectories following loss of RNA interference in Cryptococcus neoformans.bioRxiv [Preprint]. 2024 Oct 1:2024.08.15.608186. doi: 10.1101/2024.08.15.608186. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Nov 19;121(47):e2416656121. doi: 10.1073/pnas.2416656121. PMID: 39185155 Free PMC article. Updated. Preprint.
-
Distinct evolutionary trajectories following loss of RNA interference in Cryptococcus neoformans.Proc Natl Acad Sci U S A. 2024 Nov 19;121(47):e2416656121. doi: 10.1073/pnas.2416656121. Epub 2024 Nov 13. Proc Natl Acad Sci U S A. 2024. PMID: 39536081 Free PMC article.
-
Molecular typing and in vitro resistance of Cryptococcus neoformans clinical isolates obtained in Germany between 2011 and 2017.Int J Med Microbiol. 2019 Sep;309(6):151336. doi: 10.1016/j.ijmm.2019.151336. Epub 2019 Aug 16. Int J Med Microbiol. 2019. PMID: 31444102
-
Combatting the evolution of antifungal resistance in Cryptococcus neoformans.Mol Microbiol. 2020 Nov;114(5):721-734. doi: 10.1111/mmi.14565. Epub 2020 Jul 22. Mol Microbiol. 2020. PMID: 32697029 Review.
-
Antifungal drug resistance in pathogenic fungi.Med Mycol. 1998;36 Suppl 1:119-28. Med Mycol. 1998. PMID: 9988500 Review.
Cited by
-
Comparative genomics of Cryptococcus and Kwoniella reveals pathogenesis evolution and contrasting karyotype dynamics via intercentromeric recombination or chromosome fusion.bioRxiv [Preprint]. 2024 Jan 13:2023.12.27.573464. doi: 10.1101/2023.12.27.573464. bioRxiv. 2024. Update in: PLoS Biol. 2024 Jun 6;22(6):e3002682. doi: 10.1371/journal.pbio.3002682. PMID: 38234769 Free PMC article. Updated. Preprint.
-
Comparative genomics of the closely related fungal genera Cryptococcus and Kwoniella reveals karyotype dynamics and suggests evolutionary mechanisms of pathogenesis.PLoS Biol. 2024 Jun 6;22(6):e3002682. doi: 10.1371/journal.pbio.3002682. eCollection 2024 Jun. PLoS Biol. 2024. PMID: 38843310 Free PMC article.
-
Distinct evolutionary trajectories following loss of RNA interference in Cryptococcus neoformans.bioRxiv [Preprint]. 2024 Oct 1:2024.08.15.608186. doi: 10.1101/2024.08.15.608186. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Nov 19;121(47):e2416656121. doi: 10.1073/pnas.2416656121. PMID: 39185155 Free PMC article. Updated. Preprint.
-
Patterns and mechanisms of fungal genome plasticity.Curr Biol. 2025 Jun 9;35(11):R527-R544. doi: 10.1016/j.cub.2025.04.003. Curr Biol. 2025. PMID: 40494309 Review.
-
Identification of an active RNAi pathway in Candida albicans.Proc Natl Acad Sci U S A. 2024 Apr 23;121(17):e2315926121. doi: 10.1073/pnas.2315926121. Epub 2024 Apr 16. Proc Natl Acad Sci U S A. 2024. PMID: 38625945 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

