Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements
- PMID: 35918517
- PMCID: PMC9345997
- DOI: 10.1038/s42003-022-03721-1
Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements
Abstract
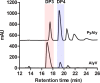
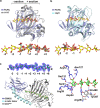
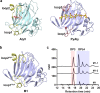
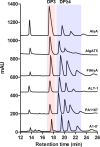
Alginate lyases can be used to produce well-defined alginate oligosaccharides (AOSs) because of their specificities for AOS products. A large number of alginate lyases have been recorded in the CAZy database; however, the majority are annotated-only alginate lyases that include little information on their products, thus limiting their applications. Here, we establish a simple and experiment-saving approach to predict product distributions for PL7 alginate lyases through extensive structural biology, bioinformatics and biochemical studies. Structural study on several PL7 alginate lyases reveals that two loops around the substrate binding cleft determine product distribution. Furthermore, a database containing the loop information of all annotated-only single-domain PL7 alginate lyases is constructed, enabling systematic exploration of the association between loop and product distribution. Based on these results, a simplified loop/product distribution relationship is proposed, giving us information on product distribution directly from the amino acid sequence.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Novel Alginate Lyase (Aly5) from a Polysaccharide-Degrading Marine Bacterium, Flammeovirga sp. Strain MY04: Effects of Module Truncation on Biochemical Characteristics, Alginate Degradation Patterns, and Oligosaccharide-Yielding Properties.Appl Environ Microbiol. 2015 Oct 30;82(1):364-74. doi: 10.1128/AEM.03022-15. Print 2016 Jan 1. Appl Environ Microbiol. 2015. PMID: 26519393 Free PMC article.
-
Structural insights into the substrate-binding cleft of AlyF reveal the first long-chain alginate-binding mode.Acta Crystallogr D Struct Biol. 2021 Mar 1;77(Pt 3):336-346. doi: 10.1107/S205979832100005X. Epub 2021 Feb 17. Acta Crystallogr D Struct Biol. 2021. PMID: 33645537
-
Characterization of a Long-Lived Alginate Lyase Derived from Shewanella Species YH1.Mar Drugs. 2017 Dec 27;16(1):4. doi: 10.3390/md16010004. Mar Drugs. 2017. PMID: 29280943 Free PMC article.
-
Structure Characteristics, Biochemical Properties, and Pharmaceutical Applications of Alginate Lyases.Mar Drugs. 2021 Nov 10;19(11):628. doi: 10.3390/md19110628. Mar Drugs. 2021. PMID: 34822499 Free PMC article. Review.
-
Alginate degrading enzymes: an updated comprehensive review of the structure, catalytic mechanism, modification method and applications of alginate lyases.Crit Rev Biotechnol. 2021 Sep;41(6):953-968. doi: 10.1080/07388551.2021.1898330. Epub 2021 May 20. Crit Rev Biotechnol. 2021. PMID: 34015998 Review.
Cited by
-
Degradation of Natural Undaria pinnatifida into Unsaturated Guluronic Acid Oligosaccharides by a Single Alginate Lyase.Mar Drugs. 2024 Oct 2;22(10):453. doi: 10.3390/md22100453. Mar Drugs. 2024. PMID: 39452861 Free PMC article.
-
Well-defined alginate oligosaccharides ameliorate joint pain and inflammation in a mouse model of gouty arthritis.Theranostics. 2024 May 19;14(8):3082-3103. doi: 10.7150/thno.95611. eCollection 2024. Theranostics. 2024. PMID: 38855180 Free PMC article.
-
Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation.Nat Commun. 2025 Mar 18;16(1):2670. doi: 10.1038/s41467-025-56754-5. Nat Commun. 2025. PMID: 40102416 Free PMC article.
-
Characterization of Multiple Alginate Lyases in a Highly Efficient Alginate-Degrading Vibrio Strain and Its Degradation Strategy.Appl Environ Microbiol. 2022 Dec 13;88(23):e0138922. doi: 10.1128/aem.01389-22. Epub 2022 Nov 21. Appl Environ Microbiol. 2022. PMID: 36409133 Free PMC article.
-
Identification and characterization of a critical loop for the high activity of alginate lyase VaAly2 from the PL7_5 subfamily.Front Microbiol. 2024 Jan 12;14:1333597. doi: 10.3389/fmicb.2023.1333597. eCollection 2023. Front Microbiol. 2024. PMID: 38282736 Free PMC article.
References
-
- Arne, et al. Uronic acid aequence in alginate from different sources. Carbohydr. Res. 1974;32:217–225. doi: 10.1016/S0008-6215(00)82100-X. - DOI
-
- Thomas F, et al. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ. Microbiol. 2012;14:2379–2394. doi: 10.1111/j.1462-2920.2012.02751.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

