Hmga2 protein loss alters nuclear envelope and 3D chromatin structure
- PMID: 35918713
- PMCID: PMC9344646
- DOI: 10.1186/s12915-022-01375-3
Hmga2 protein loss alters nuclear envelope and 3D chromatin structure
Abstract
Background: The high-mobility group Hmga family of proteins are non-histone chromatin-interacting proteins which have been associated with a number of nuclear functions, including heterochromatin formation, replication, recombination, DNA repair, transcription, and formation of enhanceosomes. Due to its role based on dynamic interaction with chromatin, Hmga2 has a pathogenic role in diverse tumors and has been mainly studied in a cancer context; however, whether Hmga2 has similar physiological functions in normal cells remains less explored. Hmga2 was additionally shown to be required during the exit of embryonic stem cells (ESCs) from the ground state of pluripotency, to allow their transition into epiblast-like cells (EpiLCs), and here, we use that system to gain further understanding of normal Hmga2 function.
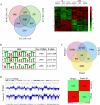
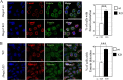
Results: We demonstrated that Hmga2 KO pluripotent stem cells fail to develop into EpiLCs. By using this experimental system, we studied the chromatin changes that take place upon the induction of EpiLCs and we observed that the loss of Hmga2 affects the histone mark H3K27me3, whose levels are higher in Hmga2 KO cells. Accordingly, a sustained expression of polycomb repressive complex 2 (PRC2), responsible for H3K27me3 deposition, was observed in KO cells. However, gene expression differences between differentiating wt vs Hmga2 KO cells did not show any significant enrichments of PRC2 targets. Similarly, endogenous Hmga2 association to chromatin in epiblast stem cells did not show any clear relationships with gene expression modification observed in Hmga2 KO. Hmga2 ChIP-seq confirmed that this protein preferentially binds to the chromatin regions associated with nuclear lamina. Starting from this observation, we demonstrated that nuclear lamina underwent severe alterations when Hmga2 KO or KD cells were induced to exit from the naïve state and this phenomenon is accompanied by a mislocalization of the heterochromatin mark H3K9me3 within the nucleus. As nuclear lamina (NL) is involved in the organization of 3D chromatin structure, we explored the possible effects of Hmga2 loss on this phenomenon. The analysis of Hi-C data in wt and Hmga2 KO cells allowed us to observe that inter-TAD (topologically associated domains) interactions in Hmga2 KO cells are different from those observed in wt cells. These differences clearly show a peculiar compartmentalization of inter-TAD interactions in chromatin regions associated or not to nuclear lamina.
Conclusions: Overall, our results indicate that Hmga2 interacts with heterochromatic lamin-associated domains, and highlight a role for Hmga2 in the crosstalk between chromatin and nuclear lamina, affecting the establishment of inter-TAD interactions.
Keywords: High mobility group proteins; Histone modifications; Lamin; Nuclear envelope; Pluripotent stem cells; Topologically associated domains.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Hmga2 is necessary for Otx2-dependent exit of embryonic stem cells from the pluripotent ground state.BMC Biol. 2016 Mar 31;14:24. doi: 10.1186/s12915-016-0246-5. BMC Biol. 2016. PMID: 27036552 Free PMC article.
-
Integrative Proteomic Profiling Reveals PRC2-Dependent Epigenetic Crosstalk Maintains Ground-State Pluripotency.Cell Stem Cell. 2019 Jan 3;24(1):123-137.e8. doi: 10.1016/j.stem.2018.10.017. Epub 2018 Nov 21. Cell Stem Cell. 2019. PMID: 30472157
-
The high mobility group protein HMGA2: a co-regulator of chromatin structure and pluripotency in stem cells?Stem Cell Rev Rep. 2009 Sep;5(3):224-30. doi: 10.1007/s12015-009-9078-9. Epub 2009 Jun 24. Stem Cell Rev Rep. 2009. PMID: 19551524 Review.
-
Polycomb-lamina antagonism partitions heterochromatin at the nuclear periphery.Nat Commun. 2022 Jul 20;13(1):4199. doi: 10.1038/s41467-022-31857-5. Nat Commun. 2022. PMID: 35859152 Free PMC article.
-
H3K27 methylation: a promiscuous repressive chromatin mark.Curr Opin Genet Dev. 2017 Apr;43:31-37. doi: 10.1016/j.gde.2016.11.001. Epub 2016 Dec 8. Curr Opin Genet Dev. 2017. PMID: 27940208 Free PMC article. Review.
Cited by
-
Decoding high mobility group A2 protein expression regulation and implications in human cancers.Discov Oncol. 2024 Jul 31;15(1):322. doi: 10.1007/s12672-024-01202-x. Discov Oncol. 2024. PMID: 39085703 Free PMC article. Review.
-
Binding to the Other Side: The AT-Hook DNA-Binding Domain Allows Nuclear Factors to Exploit the DNA Minor Groove.Int J Mol Sci. 2024 Aug 14;25(16):8863. doi: 10.3390/ijms25168863. Int J Mol Sci. 2024. PMID: 39201549 Free PMC article. Review.
-
HMGA2 promotes resistance against paclitaxel by targeting the p53 signaling pathway in colorectal cancer cells.Heliyon. 2024 May 20;10(11):e31431. doi: 10.1016/j.heliyon.2024.e31431. eCollection 2024 Jun 15. Heliyon. 2024. PMID: 38845972 Free PMC article.
-
Polymer models of chromatin organization in virally infected cells.Biochem Soc Trans. 2025 Feb 7;53(1):249-58. doi: 10.1042/BST20240598. Biochem Soc Trans. 2025. PMID: 39927819 Free PMC article. Review.
-
Engineered uterine primary myometrial cells with high-mobility group AT-hook 2 overexpression display a leiomyoma-like transcriptional and epigenomic phenotype.F S Sci. 2024 Nov;5(4):352-368. doi: 10.1016/j.xfss.2024.07.008. Epub 2024 Jul 27. F S Sci. 2024. PMID: 39074663
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

