PFKFB3-mediated glycometabolism reprogramming modulates endothelial differentiation and angiogenic capacity of placenta-derived mesenchymal stem cells
- PMID: 35918720
- PMCID: PMC9344722
- DOI: 10.1186/s13287-022-03089-3
PFKFB3-mediated glycometabolism reprogramming modulates endothelial differentiation and angiogenic capacity of placenta-derived mesenchymal stem cells
Abstract
Background: Mesenchymal stem cells (MSCs) have a great potential ability for endothelial differentiation, contributing to an effective means of therapeutic angiogenesis. Placenta-derived mesenchymal stem cells (PMSCs) have gradually attracted attention, while the endothelial differentiation has not been fully evaluated in PMSCs. Metabolism homeostasis plays an important role in stem cell differentiation, but less is known about the glycometabolic reprogramming during the PMSCs endothelial differentiation. Hence, it is critical to investigate the potential role of glycometabolism reprogramming in mediating PMSCs endothelial differentiation.
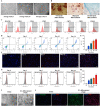
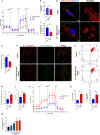
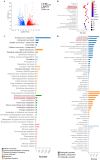
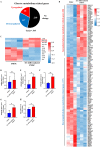
Methods: Dil-Ac-LDL uptake assay, flow cytometry, and immunofluorescence were all to verify the endothelial differentiation in PMSCs. Seahorse XF Extracellular Flux Analyzers, Mito-tracker red staining, Mitochondrial membrane potential (MMP), lactate secretion assay, and transcriptome approach were to assess the variation of mitochondrial respiration and glycolysis during the PMSCs endothelial differentiation. Glycolysis enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3) was considered a potential modulator for endothelial differentiation in PMSCs by small interfering RNA. Furthermore, transwell, in vitro Matrigel tube formation, and in vivo Matrigel plug assays were performed to evaluate the effect of PFKFB3-induced glycolysis on angiogenic capacities in this process.
Results: PMSCs possessed the superior potential of endothelial differentiation, in which the glycometabolic preference for glycolysis was confirmed. Moreover, PFKFB3-induced glycometabolism reprogramming could modulate the endothelial differentiation and angiogenic abilities of PMSCs.
Conclusions: Our results revealed that PFKFB3-mediated glycolysis is important for endothelial differentiation and angiogenesis in PMSCs. Our understanding of cellular glycometabolism and its regulatory effects on endothelial differentiation may propose and improve PMSCs as a putative strategy for clinical therapeutic angiogenesis.
Keywords: Angiogenesis; Endothelial differentiation; Glycolysis; Glycometabolism reprogramming; Mesenchymal stem cells; PFKFB3; Placenta.
© 2022. The Author(s).
Conflict of interest statement
The authors have declared that no competing interests exists.
Figures

Similar articles
-
Laminar shear stress inhibits endothelial cell metabolism via KLF2-mediated repression of PFKFB3.Arterioscler Thromb Vasc Biol. 2015 Jan;35(1):137-45. doi: 10.1161/ATVBAHA.114.304277. Epub 2014 Oct 30. Arterioscler Thromb Vasc Biol. 2015. PMID: 25359860
-
Glycometabolic reprogramming-mediated proangiogenic phenotype enhancement of cancer-associated fibroblasts in oral squamous cell carcinoma: role of PGC-1α/PFKFB3 axis.Br J Cancer. 2022 Aug;127(3):449-461. doi: 10.1038/s41416-022-01818-2. Epub 2022 Apr 20. Br J Cancer. 2022. PMID: 35444287 Free PMC article.
-
Endothelial PFKFB3 plays a critical role in angiogenesis.Arterioscler Thromb Vasc Biol. 2014 Jun;34(6):1231-9. doi: 10.1161/ATVBAHA.113.303041. Epub 2014 Apr 3. Arterioscler Thromb Vasc Biol. 2014. PMID: 24700124 Free PMC article.
-
Role of PFKFB3-driven glycolysis in sepsis.Ann Med. 2023 Dec;55(1):1278-1289. doi: 10.1080/07853890.2023.2191217. Ann Med. 2023. PMID: 37199341 Free PMC article. Review.
-
6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: A pair of valves for fine-tuning of glucose metabolism in human cancer.Mol Metab. 2019 Feb;20:1-13. doi: 10.1016/j.molmet.2018.11.013. Epub 2018 Dec 5. Mol Metab. 2019. PMID: 30553771 Free PMC article. Review.
Cited by
-
Oviduct Transcriptomic Reveals the Regulation of mRNAs and lncRNAs Related to Goat Prolificacy in the Luteal Phase.Animals (Basel). 2022 Oct 18;12(20):2823. doi: 10.3390/ani12202823. Animals (Basel). 2022. PMID: 36290212 Free PMC article.
-
Glucose Metabolism Reprogramming of Vascular Endothelial Cells and Its Implication in Development of Atherosclerosis.Rev Cardiovasc Med. 2024 Nov 22;25(11):423. doi: 10.31083/j.rcm2511423. eCollection 2024 Nov. Rev Cardiovasc Med. 2024. PMID: 39618869 Free PMC article. Review.
-
Glycometabolic Regulation of Angiogenesis: Mechanisms and Therapeutic Strategies.Int J Mol Sci. 2025 Mar 7;26(6):2386. doi: 10.3390/ijms26062386. Int J Mol Sci. 2025. PMID: 40141029 Free PMC article. Review.
-
Inflammation-Induced Alternative Splicing in Human Endothelial Cells Reveals Genetic Mechanisms of Cardiovascular Disease Risk.bioRxiv [Preprint]. 2025 Aug 2:2025.07.29.667484. doi: 10.1101/2025.07.29.667484. bioRxiv. 2025. PMID: 40766718 Free PMC article. Preprint.
References
-
- Phng LK, Belting HG. Endothelial cell mechanics and blood flow forces in vascular morphogenesis. Semin Cell Dev Biol. 2021. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

