Targeting epigenetic modifiers to reprogramme macrophages in non-resolving inflammation-driven atherosclerosis
- PMID: 35919269
- PMCID: PMC9241575
- DOI: 10.1093/ehjopen/oeab022
Targeting epigenetic modifiers to reprogramme macrophages in non-resolving inflammation-driven atherosclerosis
Erratum in
-
Corrigendum to articles in EHJ Open missing data availability statements.Eur Heart J Open. 2024 Jan 16;4(1):oead137. doi: 10.1093/ehjopen/oead137. eCollection 2024 Jan. Eur Heart J Open. 2024. PMID: 38230360 Free PMC article.
Abstract
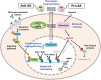
Epigenomic and epigenetic research has been providing several new insights into a variety of diseases caused by non-resolving inflammation, including cardiovascular diseases. Atherosclerosis (AS) has long been recognized as a chronic inflammatory disease of the arterial walls, characterized by local persistent and stepwise accelerating inflammation without resolution, also known as uncontrolled inflammation. The pathogenesis of AS is driven primarily by highly plastic macrophages via their polarization to pro- or anti-inflammatory phenotypes as well as other novel subtypes recently identified by single-cell sequencing. Although emerging evidence has indicated the key role of the epigenetic machinery in the regulation of macrophage plasticity, the investigation of epigenetic alterations and modifiers in AS and related inflammation is still in its infancy. An increasing number of the epigenetic modifiers (e.g. TET2, DNMT3A, HDAC3, HDAC9, JMJD3, KDM4A) have been identified in epigenetic remodelling of macrophages through DNA methylation or histone modifications (e.g. methylation, acetylation, and recently lactylation) in inflammation. These or many unexplored modifiers function to determine or switch the direction of macrophage polarization via transcriptional reprogramming of gene expression and intracellular metabolic rewiring upon microenvironmental cues, thereby representing a promising target for anti-inflammatory therapy in AS. Here, we review up-to-date findings involving the epigenetic regulation of macrophages to shed light on the mechanism of uncontrolled inflammation during AS onset and progression. We also discuss current challenges for developing an effective and safe anti-AS therapy that targets the epigenetic modifiers and propose a potential anti-inflammatory strategy that repolarizes macrophages from pro- to anti-inflammatory phenotypes.
Keywords: Atherosclerosis; Epigenetic modifier; Inflammation; Macrophage; Polarization.
© The Author(s) 2021. Published by Oxford University Press on behalf of the European Society of Cardiology.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials

