Complete mitochondrial genomes of four deep-sea echinoids: conserved mitogenome organization and new insights into the phylogeny and evolution of Echinoidea
- PMID: 35919401
- PMCID: PMC9339218
- DOI: 10.7717/peerj.13730
Complete mitochondrial genomes of four deep-sea echinoids: conserved mitogenome organization and new insights into the phylogeny and evolution of Echinoidea
Abstract
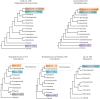
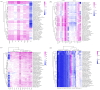
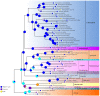
Echinoids are an important component in benthic marine environments, which occur at all depths from the shallow-water hard substrates to abyssal depths. To date, the phylogeny of the sea urchins and the macro-evolutionary processes of deep-sea and shallow water groups have not yet been fully resolved. In the present study, we sequenced the complete mitochondrial genomes (mitogenomes) of four deep-sea sea urchins (Echinoidea), which were the first representatives of the orders Aspidodiadematoida, Pedinoida and Echinothurioida, respectively. The gene content and arrangement were highly conserved in echinoid mitogenomes. The tRNA-Ser AGY with DHU arm was detected in the newly sequenced echinoid mitogenomes, representing an ancestral structure of tRNA-Ser AGY. No difference was found between deep-sea and shallow water groups in terms of base composition and codon usage. The phylogenetic analysis showed that all the orders except Spatangoida were monophyletic. The basal position of Cidaroida was supported. The closest relationship of Scutelloida and Echinolampadoida was confirmed. Our phylogenetic analysis shed new light on the position of Arbacioida, which supported that Arbacioida was most related with the irregular sea urchins instead of Stomopneustoida. The position Aspidodiadematoida (((Aspidodiadematoida + Pedinoida) + Echinothurioida) + Diadematoida) revealed by mitogenomic data discredited the hypothesis based on morphological evidences. The macro-evolutionary pattern revealed no simple onshore-offshore or an opposite hypothesis. But the basal position of the deep-sea lineages indicated the important role of deep sea in generating the current diversity of the class Echinoidea.
Keywords: Deep-sea; Echinoidea; Evolution; Mitochondrial genome; Phylogenetic relationship.
©2022 Sun et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



Similar articles
-
The mitochondrial genome of the deep-sea pyramid urchin Echinocrepis rostrata (Echinoidea: Holasteroida: Pourtalesiidae).Mitochondrial DNA B Resour. 2024 Mar 23;9(3):390-393. doi: 10.1080/23802359.2024.2333572. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 38529110 Free PMC article.
-
A combined morphological and molecular phylogeny for sea urchins (Echinoidea: Echinodermata).Philos Trans R Soc Lond B Biol Sci. 1995 Jan 30;347(1320):213-34. doi: 10.1098/rstb.1995.0023. Philos Trans R Soc Lond B Biol Sci. 1995. PMID: 7746863 Review.
-
The first complete mitochondrial genome of the sand dollar Sinaechinocyamus mai (Echinoidea: Clypeasteroida).Genomics. 2020 Mar;112(2):1686-1693. doi: 10.1016/j.ygeno.2019.10.007. Epub 2019 Oct 17. Genomics. 2020. PMID: 31629878 Free PMC article.
-
A phylogenomic resolution of the sea urchin tree of life.BMC Evol Biol. 2018 Dec 13;18(1):189. doi: 10.1186/s12862-018-1300-4. BMC Evol Biol. 2018. PMID: 30545284 Free PMC article.
-
Mind the gap! The mitochondrial control region and its power as a phylogenetic marker in echinoids.BMC Evol Biol. 2018 May 30;18(1):80. doi: 10.1186/s12862-018-1198-x. BMC Evol Biol. 2018. PMID: 29848319 Free PMC article.
Cited by
-
A novel deep-benthic sea cucumber species of Benthodytes (Holothuroidea, Elasipodida, Psychropotidae) and its comprehensive mitochondrial genome sequencing and evolutionary analysis.BMC Genomics. 2024 Jul 13;25(1):689. doi: 10.1186/s12864-024-10607-5. BMC Genomics. 2024. PMID: 39003448 Free PMC article.
-
The mitochondrial genome of the deep-sea pyramid urchin Echinocrepis rostrata (Echinoidea: Holasteroida: Pourtalesiidae).Mitochondrial DNA B Resour. 2024 Mar 23;9(3):390-393. doi: 10.1080/23802359.2024.2333572. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 38529110 Free PMC article.
-
A 104-Ma record of deep-sea Atelostomata (Holasterioda, Spatangoida, irregular echinoids) - a story of persistence, food availability and a big bang.PLoS One. 2023 Aug 9;18(8):e0288046. doi: 10.1371/journal.pone.0288046. eCollection 2023. PLoS One. 2023. PMID: 37556403 Free PMC article.
-
Chromosome-level genome assembly and annotation of the black sea urchin Arbacia lixula (Linnaeus, 1758).DNA Res. 2024 Aug 1;31(4):dsae020. doi: 10.1093/dnares/dsae020. DNA Res. 2024. PMID: 38908014 Free PMC article.
-
Embracing the taxonomic and topological stability of phylogenomics.Sci Rep. 2024 Feb 19;14(1):4088. doi: 10.1038/s41598-024-54208-4. Sci Rep. 2024. PMID: 38374111 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

