Evolution of ACE2-independent SARS-CoV-2 infection and mouse adaption after passage in cells expressing human and mouse ACE2
- PMID: 35919871
- PMCID: PMC9338707
- DOI: 10.1093/ve/veac063
Evolution of ACE2-independent SARS-CoV-2 infection and mouse adaption after passage in cells expressing human and mouse ACE2
Abstract
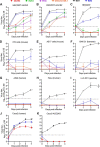
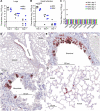
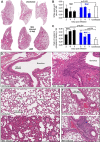
Human ACE2 Human angiotensin converting enzyme 2 (hACE2) is the key cell attachment and entry receptor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), with the original SARS-CoV-2 isolates unable to use mouse ACE2 (mACE2). Herein we describe the emergence of a SARS-CoV-2 strain capable of ACE2-independent infection and the evolution of mouse-adapted (MA) SARS-CoV-2 by in vitro serial passaging of virus in co-cultures of cell lines expressing hACE2 and mACE2. MA viruses evolved with up to five amino acid changes in the spike protein, all of which have been seen in human isolates. MA viruses replicated to high titers in C57BL/6J mouse lungs and nasal turbinates and caused characteristic lung histopathology. One MA virus also evolved to replicate efficiently in several ACE2-negative cell lines across several species, including clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) ACE2 knockout cells. An E484D substitution is likely involved in ACE2-independent entry and has appeared in only ≈0.003 per cent of human isolates globally, suggesting that it provided no significant selection advantage in humans. ACE2-independent entry reveals a SARS-CoV-2 infection mechanism that has potential implications for disease pathogenesis, evolution, tropism, and perhaps also intervention development.
Keywords: ACE2-independent infection; COVID-19; SARS-CoV-2; mouse adaptation.
© The Author(s) 2022. Published by Oxford University Press.
Figures



References
-
- Andrews S. (2010) ‘A Quality Control Tool for High Throughput Sequence Data’. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed 21 July 2022.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

