Epidemiological investigation and phylogenetic analysis of Classical Swine Fever virus in Yunnan province from 2015 to 2021
- PMID: 35920121
- PMCID: PMC9346530
- DOI: 10.4142/jvs.22042
Epidemiological investigation and phylogenetic analysis of Classical Swine Fever virus in Yunnan province from 2015 to 2021
Abstract
Background: Classical swine fever virus (CSFV), the causative agent of classical swine fever (CFS), is a highly contagious disease that poses a serious threat to Chinese pig populations.
Objectives: Many provinces of China, such as Shandong, Henan, Hebei, Heilongjiang, and Liaoning provinces, have reported epidemics of CSFV, while the references to the epidemic of CSFV in Yunnan province are rare. This study examined the epidemic characteristics of the CSFV in Yunnan province.
Methods: In this study, 326 tissue samples were collected from different regions in Yunnan province from 2015 to 2021. A reverse transcription-polymerase chain reaction (RT-PCR), sequences analysis, and phylogenetic analysis were performed for the pathogenic detection and analysis of these 326 clinical specimens.
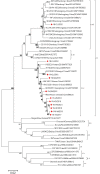
Results: Approximately 3.37% (11/326) of specimens tested positive for the CSFV by RT-PCR, which is lower than that of other regions of China. Sequence analysis of the partial E2 sequences of eleven CSFV strains showed that they shared 89.0-100.0% nucleotide (nt) and 95.0-100.0% amino acid (aa) homology, respectively. Phylogenetic analysis showed that these novel isolates belonged to the subgenotypes 2.1c and 2.1d, with subgenotype 2.1c being predominant.
Conclusions: The CSFV was sporadic in China's Yunnan province from 2015 to 2021. Both 2.1c and 2.1d subgenotypes were found in this region, but 2.1c was dominant.
Keywords: Classical swine fever virus; Yunnan province; sequence analysis; subgenotype 2.1c.
© 2022 The Korean Society of Veterinary Science.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Genetic diversity of subgenotype 2.1 isolates of classical swine fever virus.Infect Genet Evol. 2016 Jul;41:218-226. doi: 10.1016/j.meegid.2016.04.002. Epub 2016 Apr 13. Infect Genet Evol. 2016. PMID: 27085291
-
Phylogenetic analysis using E2 gene of classical swine fever virus reveals a new subgenotype in China.Infect Genet Evol. 2013 Jul;17:231-8. doi: 10.1016/j.meegid.2013.04.004. Epub 2013 Apr 19. Infect Genet Evol. 2013. PMID: 23608662
-
Sub-subgenotype 2.1c isolates of classical swine fever virus are dominant in Guangdong province of China, 2018.Infect Genet Evol. 2019 Mar;68:212-217. doi: 10.1016/j.meegid.2018.12.029. Epub 2018 Dec 25. Infect Genet Evol. 2019. PMID: 30592978
-
Laboratory diagnosis, epizootiology, and efficacy of marker vaccines in classical swine fever: a review.Vet Q. 2000 Oct;22(4):182-8. doi: 10.1080/01652176.2000.9695054. Vet Q. 2000. PMID: 11087126 Review.
-
Classical swine fever in China: a minireview.Vet Microbiol. 2014 Aug 6;172(1-2):1-6. doi: 10.1016/j.vetmic.2014.04.004. Epub 2014 Apr 13. Vet Microbiol. 2014. PMID: 24793098 Review.
Cited by
-
Development of a quadruplex real-time quantitative RT-PCR for detection and differentiation of PHEV, PRV, CSFV, and JEV.Front Vet Sci. 2023 Oct 31;10:1276505. doi: 10.3389/fvets.2023.1276505. eCollection 2023. Front Vet Sci. 2023. PMID: 38026635 Free PMC article.
-
Classical Swine Fever Virus Envelope Glycoproteins Erns, E1, and E2 Activate IL-10-STAT1-MX1/OAS1 Antiviral Pathway via Replacing Classical IFNα/β.Biomolecules. 2025 Jan 31;15(2):200. doi: 10.3390/biom15020200. Biomolecules. 2025. PMID: 40001503 Free PMC article.
-
Development of a triplex crystal digital RT-PCR for the detection of PHEV, PRV, and CSFV.Front Vet Sci. 2024 Dec 12;11:1462880. doi: 10.3389/fvets.2024.1462880. eCollection 2024. Front Vet Sci. 2024. PMID: 39726583 Free PMC article.
-
The regulation of cell homeostasis and antiviral innate immunity by autophagy during classical swine fever virus infection.Emerg Microbes Infect. 2023 Dec;12(1):2164217. doi: 10.1080/22221751.2022.2164217. Emerg Microbes Infect. 2023. PMID: 36583373 Free PMC article.
References
-
- Becher P, Avalos Ramirez R, Orlich M, Cedillo Rosales S, König M, Schweizer M, et al. Genetic and antigenic characterization of novel pestivirus genotypes: implications for classification. Virology. 2003;311(1):96–104. - PubMed
-
- Ji W, Guo Z, Ding NZ, He CQ. Studying classical swine fever virus: making the best of a bad virus. Virus Res. 2015;197:35–47. - PubMed
-
- Luo Y, Ji S, Liu Y, Lei JL, Xia SL, Wang Y, et al. Isolation and characterization of a moderately virulent classical swine fever virus emerging in China. Transbound Emerg Dis. 2017;64(6):1848–1857. - PubMed
-
- Luo TR, Liao SH, Wu XS, Feng L, Yuan ZX, Li H, et al. Phylogenetic analysis of the E2 gene of classical swine fever virus from the Guangxi Province of southern China. Virus Genes. 2011;42(3):347–354. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

