Identifying group-specific primers for environmental Heterolobosa by high-throughput sequencing
- PMID: 35920130
- PMCID: PMC9437880
- DOI: 10.1111/1751-7915.14080
Identifying group-specific primers for environmental Heterolobosa by high-throughput sequencing
Abstract
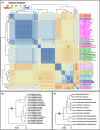
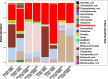
Diversity of Heterolobosea (Excavata) in environments is poorly understood despite their ecological occurrence and health-associated risk, partly because this group tends to be under-covered by most universal eukaryotic primers used for sequencing. To overcome the limits of the traditional morpho-taxonomy-based biomonitoring, we constructed a primer database listing existing and newly designed specific primer pairs that have been evaluated for Heterolobosea 18S rRNA sequencing. In silico taxonomy performance against the current SILVA SSU database allowed the selection of primer pairs that were next evaluated on reference culture amoebal strains. Two primer pairs were retained for monitoring the diversity of Heterolobosea in freshwater environments, using high-throughput sequencing. Results showed that one of the newly designed primer pairs allowed species-level identification of most heterolobosean sequences. Such primer pair could enable informative, cultivation-free assays for characterizing heterolobosean populations in various environments.
© 2022 The Authors. Microbial Biotechnology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

