CRISPR/Cas-based tools for the targeted control of plant viruses
- PMID: 35920132
- PMCID: PMC9562834
- DOI: 10.1111/mpp.13252
CRISPR/Cas-based tools for the targeted control of plant viruses
Abstract
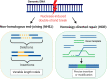
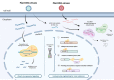
Plant viruses are known to infect most economically important crops and pose a major threat to global food security. Currently, few resistant host phenotypes have been delineated, and while chemicals are used for crop protection against insect pests and bacterial or fungal diseases, these are inefficient against viral diseases. Genetic engineering emerged as a way of modifying the plant genome by introducing functional genes in plants to improve crop productivity under adverse environmental conditions. Recently, new breeding technologies, and in particular the exciting CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins) technology, was shown to be a powerful alternative to engineer resistance against plant viruses, thus has great potential for reducing crop losses and improving plant productivity to directly contribute to food security. Indeed, it could circumvent the "Genetic modification" issues because it allows for genome editing without the integration of foreign DNA or RNA into the genome of the host plant, and it is simpler and more versatile than other new breeding technologies. In this review, we describe the predominant features of the major CRISPR/Cas systems and outline strategies for the delivery of CRISPR/Cas reagents to plant cells. We also provide an overview of recent advances that have engineered CRISPR/Cas-based resistance against DNA and RNA viruses in plants through the targeted manipulation of either the viral genome or susceptibility factors of the host plant genome. Finally, we provide insight into the limitations and challenges that CRISPR/Cas technology currently faces and discuss a few alternative applications of the technology in virus research.
Keywords: CRISPR/Cas; crop; genome editing; plant viruses.
© 2022 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures

Similar articles
-
CRISPR/Cas systems versus plant viruses: engineering plant immunity and beyond.Plant Physiol. 2021 Aug 3;186(4):1770-1785. doi: 10.1093/plphys/kiab220. Plant Physiol. 2021. PMID: 35237805 Free PMC article.
-
Plant Viruses: From Targets to Tools for CRISPR.Viruses. 2021 Jan 19;13(1):141. doi: 10.3390/v13010141. Viruses. 2021. PMID: 33478128 Free PMC article. Review.
-
Tools and targets: The dual role of plant viruses in CRISPR-Cas genome editing.Plant Genome. 2023 Jun;16(2):e20220. doi: 10.1002/tpg2.20220. Epub 2022 Jun 14. Plant Genome. 2023. PMID: 35698891 Review.
-
Tweaking genome-editing approaches for virus interference in crop plants.Plant Physiol Biochem. 2020 Feb;147:242-250. doi: 10.1016/j.plaphy.2019.12.022. Epub 2019 Dec 19. Plant Physiol Biochem. 2020. PMID: 31881433 Review.
-
CRISPR/Cas: A powerful tool for gene function study and crop improvement.J Adv Res. 2020 Oct 21;29:207-221. doi: 10.1016/j.jare.2020.10.003. eCollection 2021 Mar. J Adv Res. 2020. PMID: 33842017 Free PMC article. Review.
Cited by
-
Role of Plant Virus Movement Proteins in Suppression of Host RNAi Defense.Int J Mol Sci. 2023 May 20;24(10):9049. doi: 10.3390/ijms24109049. Int J Mol Sci. 2023. PMID: 37240394 Free PMC article. Review.
-
Rapid detection of avian leukemia virus using CRISPR/Cas13a based lateral flow dipstick.Front Vet Sci. 2024 Aug 16;11:1424238. doi: 10.3389/fvets.2024.1424238. eCollection 2024. Front Vet Sci. 2024. PMID: 39220765 Free PMC article.
-
Plant protection from virus: a review of different approaches.Front Plant Sci. 2023 Jun 12;14:1163270. doi: 10.3389/fpls.2023.1163270. eCollection 2023. Front Plant Sci. 2023. PMID: 37377807 Free PMC article. Review.
-
CRISPR/Cas9-Mediated Resistance to Wheat Dwarf Virus in Hexaploid Wheat (Triticum aestivum L.).Viruses. 2024 Aug 29;16(9):1382. doi: 10.3390/v16091382. Viruses. 2024. PMID: 39339858 Free PMC article.
-
Plant translational reprogramming for stress resilience.Front Plant Sci. 2023 Feb 24;14:1151587. doi: 10.3389/fpls.2023.1151587. eCollection 2023. Front Plant Sci. 2023. PMID: 36909402 Free PMC article. Review.
References
-
- Ali, Z. , Abul‐faraj, A. , Li, L. , Ghosh, N. , Piatek, M. , Mahjoub, A. et al. (2015) Efficient virus‐mediated genome editing in plants using the CRISPR/Cas9 system. Molecular Plant, 8, 1288–1291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

