Phylogenomic Evidence for the Origin of Obligate Anaerobic Anammox Bacteria Around the Great Oxidation Event
- PMID: 35920138
- PMCID: PMC9387917
- DOI: 10.1093/molbev/msac170
Phylogenomic Evidence for the Origin of Obligate Anaerobic Anammox Bacteria Around the Great Oxidation Event
Abstract
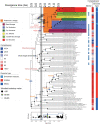
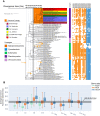
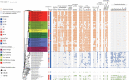
The anaerobic ammonium oxidation (anammox) bacteria can transform ammonium and nitrite to dinitrogen gas, and this obligate anaerobic process accounts for up to half of the global nitrogen loss in surface environments. Yet its origin and evolution, which may give important insights into the biogeochemistry of early Earth, remain enigmatic. Here, we performed a comprehensive phylogenomic and molecular clock analysis of anammox bacteria within the phylum Planctomycetes. After accommodating the uncertainties and factors influencing time estimates, which include implementing both a traditional cyanobacteria-based and a recently developed mitochondria-based molecular dating approach, we estimated a consistent origin of anammox bacteria at early Proterozoic and most likely around the so-called Great Oxidation Event (GOE; 2.32-2.5 Ga) which fundamentally changed global biogeochemical cycles. We further showed that during the origin of anammox bacteria, genes involved in oxidative stress adaptation, bioenergetics, and anammox granules formation were recruited, which might have contributed to their survival on an increasingly oxic Earth. Our findings suggest the rising levels of atmospheric oxygen, which made nitrite increasingly available, was a potential driving force for the emergence of anammox bacteria. This is one of the first studies that link the GOE to the evolution of obligate anaerobic bacteria.
Keywords: anammox bacteria; molecular dating analysis; planctomycetes.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures
References
-
- Ader M, Sansjofre P, Halverson GP, Busigny V, Trindade RIF, Kunzmann M, Nogueira ACR. 2014. Ocean redox structure across the Late Neoproterozoic Oxygenation Event: a nitrogen isotope perspective. Earth Planet Sci Lett. 396:1–13.
-
- Ader M, Thomazo C, Sansjofre P, Busigny V, Papineau D, Laffont R, Cartigny P, Halverson GP. 2016. Interpretation of the nitrogen isotopic composition of Precambrian sedimentary rocks: assumptions and perspectives. Chem Geol. 429:93–110.
-
- Anbar AD, Duan Y, Lyons TW, Arnold GL, Kendall B, Creaser RA, Kaufman AJ, Gordon GW, Scott C, Garvin J, et al. 2007. A whiff of oxygen before the Great Oxidation Event? Science 317:1903–1906. - PubMed
-
- Battistuzzi FU, Hedges SB. 2009. A Major Clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol. 26:335–343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

