FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations
- PMID: 35920317
- PMCID: PMC9410875
- DOI: 10.1093/nar/gkac673
FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations
Abstract
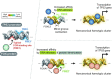
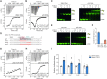
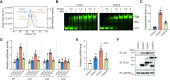
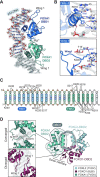
Although both the p53 and forkhead box (FOX) family proteins are key transcription factors associated with cancer progression, their direct relationship is unknown. Here, we found that FOX family proteins bind to the non-canonical homotypic cluster of the p53 promoter region (TP53). Analysis of crystal structures of FOX proteins (FOXL2 and FOXA1) bound to the p53 homotypic cluster indicated that they interact with a 2:1 stoichiometry accommodated by FOX-induced DNA allostery. In particular, FOX proteins exhibited distinct dimerization patterns in recognition of the same p53-DNA; dimer formation of FOXA1 involved protein-protein interaction, but FOXL2 did not. Biochemical and biological functional analyses confirmed the cooperative binding of FOX proteins to the TP53 promoter for the transcriptional activation of TP53. In addition, up-regulation of TP53 was necessary for FOX proteins to exhibit anti-proliferative activity in cancer cells. These analyses reveal the presence of a discrete characteristic within FOX family proteins in which FOX proteins regulate the transcription activity of the p53 tumor suppressor via cooperative binding to the TP53 promoter in alternative dimer configurations.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
The expression of p21 is upregulated by forkhead box A1/2 in p53-null H1299 cells.FEBS Lett. 2014 Nov 3;588(21):4065-70. doi: 10.1016/j.febslet.2014.09.033. Epub 2014 Oct 2. FEBS Lett. 2014. PMID: 25281925
-
Genetic features of TP53 mutation and its downstream FOXA1 in prostate cancer.Biosci Trends. 2022 Jul 20;16(3):221-229. doi: 10.5582/bst.2022.01235. Epub 2022 Jun 28. Biosci Trends. 2022. PMID: 35768267
-
NFI transcription factors interact with FOXA1 to regulate prostate-specific gene expression.Mol Endocrinol. 2014 Jun;28(6):949-64. doi: 10.1210/me.2013-1213. Epub 2014 May 6. Mol Endocrinol. 2014. PMID: 24801505 Free PMC article.
-
Modulation of androgen receptor by FOXA1 and FOXO1 factors in prostate cancer.Int J Biol Sci. 2014 Jun 5;10(6):614-9. doi: 10.7150/ijbs.8389. eCollection 2014. Int J Biol Sci. 2014. PMID: 24948874 Free PMC article. Review.
-
Human FOX gene family (Review).Int J Oncol. 2004 Nov;25(5):1495-500. Int J Oncol. 2004. PMID: 15492844 Review.
Cited by
-
The disordered p53 transactivation domain is the target of FOXO4 and the senolytic compound FOXO4-DRI.Nat Commun. 2025 Jul 1;16(1):5672. doi: 10.1038/s41467-025-60844-9. Nat Commun. 2025. PMID: 40593617 Free PMC article.
-
Structurally Oriented Classification of FOXA1 Alterations Identifies Prostate Cancers with Opposing Clinical Outcomes and Distinct Molecular and Immunologic Subtypes.Clin Cancer Res. 2025 Mar 3;31(5):936-948. doi: 10.1158/1078-0432.CCR-24-3471. Clin Cancer Res. 2025. PMID: 39745364 Free PMC article.
-
Gain-of-Function Chromatin Remodeling Activity of Oncogenic FOXL2C134W Reprograms Glucocorticoid Receptor Occupancy to Drive Granulosa Cell Tumors.Cancer Res. 2025 Mar 3;85(5):875-893. doi: 10.1158/0008-5472.CAN-24-2341. Cancer Res. 2025. PMID: 39652611 Free PMC article.
-
Footprint-C reveals transcription factor modes in local clusters and long-range chromatin interactions.Nat Commun. 2024 Dec 30;15(1):10922. doi: 10.1038/s41467-024-55403-7. Nat Commun. 2024. PMID: 39738122 Free PMC article.
-
Conserved methylation signatures associate with the tumor immune microenvironment and immunotherapy response.Genome Med. 2024 Apr 2;16(1):47. doi: 10.1186/s13073-024-01318-3. Genome Med. 2024. PMID: 38566132 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

