Robust counterselection and advanced λRed recombineering enable markerless chromosomal integration of large heterologous constructs
- PMID: 35920321
- PMCID: PMC9410887
- DOI: 10.1093/nar/gkac649
Robust counterselection and advanced λRed recombineering enable markerless chromosomal integration of large heterologous constructs
Abstract
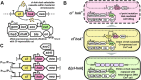
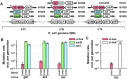
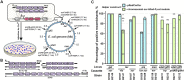
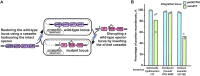
Despite advances in bacterial genome engineering, delivery of large synthetic constructs remains challenging in practice. In this study, we propose a straightforward and robust approach for the markerless integration of DNA fragments encoding whole metabolic pathways into the genome. This approach relies on the replacement of a counterselection marker with cargo DNA cassettes via λRed recombineering. We employed a counterselection strategy involving a genetic circuit based on the CI repressor of λ phage. Our design ensures elimination of most spontaneous mutants, and thus provides a counterselection stringency close to the maximum possible. We improved the efficiency of integrating long PCR-generated cassettes by exploiting the Ocr antirestriction function of T7 phage, which completely prevents degradation of unmethylated DNA by restriction endonucleases in wild-type bacteria. The employment of highly restrictive counterselection and ocr-assisted λRed recombineering allowed markerless integration of operon-sized cassettes into arbitrary genomic loci of four enterobacterial species with an efficiency of 50-100%. In the case of Escherichia coli, our strategy ensures simple combination of markerless mutations in a single strain via P1 transduction. Overall, the proposed approach can serve as a general tool for synthetic biology and metabolic engineering in a range of bacterial hosts.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Court D.L., Sawitzke J.A., Thomason L.C.. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002; 36:361–388. - PubMed
-
- Datta S., Costantino N., Court D.L.. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006; 379:109–115. - PubMed
-
- Murphy K.C., Campellone K.G., Poteete A.R.. PCR-mediated gene replacement in Escherichia coli. Gene. 2000; 246:321–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

