The Fluorescent Enzyme Cascade Detects Low Abundance Protein Modifications Suitable for the Assembly of Functionally Annotated Modificatome Databases
- PMID: 35920326
- PMCID: PMC9805100
- DOI: 10.1002/cbic.202200399
The Fluorescent Enzyme Cascade Detects Low Abundance Protein Modifications Suitable for the Assembly of Functionally Annotated Modificatome Databases
Abstract
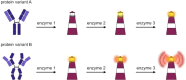
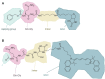
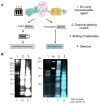
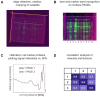
Pathophysiological functions of proteins critically depend on both their chemical composition, including post-translational modifications, and their three-dimensional structure, commonly referred to as structure-activity relationship. Current analytical methods, like capillary electrophoresis or mass spectrometry, suffer from limitations, such as the detection of unexpected modifications at low abundance and their insensitivity to conformational changes. Building on previous enzyme-based analytical methods, we here introduce a fluorescence-based enzyme cascade (fEC), which can detect diverse chemical and conformational variations in protein samples and assemble them into digital databases. Together with complementary analytical methods an automated fEC analysis established unique modification-function relationships, which can be expanded to a proteome-wide scale, i. e. a functionally annotated modificatome. The fEC offers diverse applications, including hypersensitive biomarker detection in complex samples.
Keywords: analytical methods; conformation analysis; enzymes; functional annotation; protein modifications.
© 2022 The Authors. ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
Hans Brandstetter holds a patent on the enzyme cascade based detection method (US patent 10,775,384).
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

