Precise epigenomic editing with a SunTag-based modular epigenetic toolkit
- PMID: 35920441
- PMCID: PMC9665145
- DOI: 10.1080/15592294.2022.2106646
Precise epigenomic editing with a SunTag-based modular epigenetic toolkit
Abstract
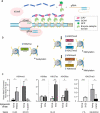
Epigenetic regulation is a crucial factor controlling gene expression. Here, we report our CRISPR/dCas9-based modular epigenetic toolkit that enables gene-specific modulation of epigenetic architecture. By modifying the SunTag framework of dCas9 tagged with five GCN4 moieties, each epigenetic writer is bound to scFv and target-specific sgRNA, and this system is able to modify multiple epigenetic marks in a target-specific manner. We successfully demonstrated that this system is efficient in modifying individual histone post-translational modifications. We display its utility as a tool to understand the contributions of specific histone marks on gene expression by screening a large promoter region and identifying differential outcomes with high base-pair resolution. This epigenetic toolkit can be easily altered with a large variety of epigenetic effectors and is a useful tool for researchers to use in understanding gene-specific epigenetic changes and their relation to gene expression.
Keywords: CRISPR; Epigenetics; epigenetic writers; epigenome editing; histone post-translational modifications.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
-
- Holtzman L, Gersbach CA.. Editing the epigenome: reshaping the genomic landscape. Annu Rev Genomics Hum Genet. 2018;19:43–71. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
