Hematopoietic plasticity mapped in Drosophila and other insects
- PMID: 35920811
- PMCID: PMC9348853
- DOI: 10.7554/eLife.78906
Hematopoietic plasticity mapped in Drosophila and other insects
Abstract
Hemocytes, similar to vertebrate blood cells, play important roles in insect development and immunity, but it is not well understood how they perform their tasks. New technology, in particular single-cell transcriptomic analysis in combination with Drosophila genetics, may now change this picture. This review aims to make sense of recently published data, focusing on Drosophila melanogaster and comparing to data from other drosophilids, the malaria mosquito, Anopheles gambiae, and the silkworm, Bombyx mori. Basically, the new data support the presence of a few major classes of hemocytes: (1) a highly heterogenous and plastic class of professional phagocytes with many functions, called plasmatocytes in Drosophila and granular cells in other insects. (2) A conserved class of cells that control melanin deposition around parasites and wounds, called crystal cells in D. melanogaster, and oenocytoids in other insects. (3) A new class of cells, the primocytes, so far only identified in D. melanogaster. They are related to cells of the so-called posterior signaling center of the larval hematopoietic organ, which controls the hematopoiesis of other hemocytes. (4) Different kinds of specialized cells, like the lamellocytes in D. melanogaster, for the encapsulation of parasites. These cells undergo rapid evolution, and the homology relationships between such cells in different insects are uncertain. Lists of genes expressed in the different hemocyte classes now provide a solid ground for further investigation of function.
Keywords: Drosophila; cell biology; chromosomes; gene expression; hematopoiesis; hemocytes; immunity; lepidoptera; mosquitoes.
© 2022, Hultmark and Andó.
Conflict of interest statement
DH, IA No competing interests declared
Figures


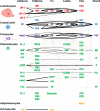



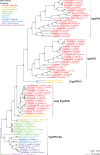

Similar articles
-
Hemocyte Clusters Defined by scRNA-Seq in Bombyx mori: In Silico Analysis of Predicted Marker Genes and Implications for Potential Functional Roles.Front Immunol. 2022 Feb 25;13:852702. doi: 10.3389/fimmu.2022.852702. eCollection 2022. Front Immunol. 2022. PMID: 35281044 Free PMC article.
-
Two hemocyte lineages exist in silkworm larval hematopoietic organ.PLoS One. 2010 Jul 28;5(7):e11816. doi: 10.1371/journal.pone.0011816. PLoS One. 2010. PMID: 20676370 Free PMC article.
-
Hemocyte differentiation in the hematopoietic organs of the silkworm, Bombyx mori: prohemocytes have the function of phagocytosis.Cell Tissue Res. 2005 Jun;320(3):535-43. doi: 10.1007/s00441-004-1038-8. Epub 2005 Apr 22. Cell Tissue Res. 2005. PMID: 15846518
-
Drosophila hemopoiesis and cellular immunity.J Immunol. 2007 Apr 15;178(8):4711-6. doi: 10.4049/jimmunol.178.8.4711. J Immunol. 2007. PMID: 17404248 Review.
-
Insect hemocytes and their role in immunity.Insect Biochem Mol Biol. 2002 Oct;32(10):1295-309. doi: 10.1016/s0965-1748(02)00092-9. Insect Biochem Mol Biol. 2002. PMID: 12225920 Review.
Cited by
-
Drosophila melanogaster as a model to study innate immune memory.Front Microbiol. 2022 Oct 20;13:991678. doi: 10.3389/fmicb.2022.991678. eCollection 2022. Front Microbiol. 2022. PMID: 36338030 Free PMC article. Review.
-
Transdifferentiation of plasmatocytes to crystal cells in the lymph gland of Drosophila melanogaster.EMBO Rep. 2025 Apr;26(8):2077-2097. doi: 10.1038/s44319-025-00366-z. Epub 2025 Mar 12. EMBO Rep. 2025. PMID: 40075235 Free PMC article.
-
Drosophila are hosts to the first described parasitoid wasp of adult flies.Nature. 2024 Sep;633(8031):840-847. doi: 10.1038/s41586-024-07919-7. Epub 2024 Sep 11. Nature. 2024. PMID: 39261731 Free PMC article.
-
Distinctive features of Zaprionus indianus hemocyte differentiation and function revealed by transcriptomic analysis.Front Immunol. 2023 Dec 21;14:1322381. doi: 10.3389/fimmu.2023.1322381. eCollection 2023. Front Immunol. 2023. PMID: 38187383 Free PMC article.
-
Insect-pathogen crosstalk and the cellular-molecular mechanisms of insect immunity: uncovering the underlying signaling pathways and immune regulatory function of non-coding RNAs.Front Immunol. 2023 Aug 24;14:1169152. doi: 10.3389/fimmu.2023.1169152. eCollection 2023. Front Immunol. 2023. PMID: 37691928 Free PMC article. Review.
References
-
- Avet-Rochex A, Boyer K, Polesello C, Gobert V, Osman D, Roch F, Augé B, Zanet J, Haenlin M, Waltzer L. An in vivo RNA interference screen identifies gene networks controlling Drosophila melanogaster blood cell homeostasis. BMC Developmental Biology. 2010;10:65. doi: 10.1186/1471-213X-10-65. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

