Identification of a Golgi-localized peptide reveals a minimal Golgi-targeting motif
- PMID: 35921174
- PMCID: PMC9635300
- DOI: 10.1091/mbc.E22-03-0091
Identification of a Golgi-localized peptide reveals a minimal Golgi-targeting motif
Abstract
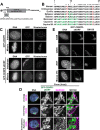
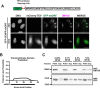
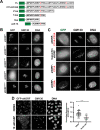
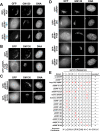
Prior work has identified signal sequences and motifs that are necessary and sufficient to target proteins to specific subcellular regions and organelles such as the plasma membrane, nucleus, endoplasmic reticulum, and mitochondria. In contrast, minimal sequence motifs that are sufficient for Golgi localization remain largely elusive. In this work, we identified a 37-amino acid alternative open reading frame (altORF) within the mRNA of the centromere protein CENP-R. This altORF peptide localizes specifically to the cytoplasmic surface of the Golgi apparatus. Through mutational analysis, we identify a minimal 10-amino acid sequence and a critical cysteine residue that are necessary and sufficient for Golgi localization. Pharmacological perturbations suggest that this peptide undergoes lipid modification to promote its localization. Together, our work defines a minimal sequence that is sufficient for Golgi targeting and provide a valuable Golgi marker for live cell imaging.
Figures

References
-
- Aicart-Ramos C, Valero RA, Rodriguez-Crespo I (2011). Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta 1808, 2981–2994. - PubMed
-
- Cheeseman IM, Desai A (2005). A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE 2005, pl1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

