Mitophagy initiates retrograde mitochondrial-nuclear signaling to guide retinal pigment cell heterogeneity
- PMID: 35921555
- PMCID: PMC9980637
- DOI: 10.1080/15548627.2022.2109286
Mitophagy initiates retrograde mitochondrial-nuclear signaling to guide retinal pigment cell heterogeneity
Abstract
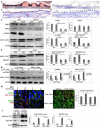
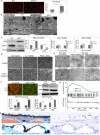
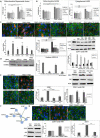
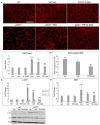
Age-related macular degeneration (AMD), the leading cause of blindness among the elderly, is without treatment for early disease. Degenerative retinal pigment epithelial (RPE) cell heterogeneity is a well-recognized but understudied pathogenic factor. Due to the daily phagocytosis of photoreceptor outer segments, unique photo-oxidative stress, and high metabolism for maintaining vision, the RPE has robust macroautophagy/autophagy, and mitochondrial and antioxidant networks. However, the autophagy subtype, mitophagy, in the RPE and AMD is understudied. Here, we found decreased PINK1 (PTEN induced kinase 1) in perifoveal RPE of early AMD eyes. PINK1-deficient RPE have impaired mitophagy and mitochondrial function that triggers death-resistant epithelial-mesenchymal transition (EMT). This reprogramming is mediated by novel retrograde mitochondrial-nuclear signaling (RMNS) through superoxide, NFE2L2 (NFE2 like bZIP transcription factor 2), TXNRD1 (thioredoxin reductase 1), and phosphoinositide 3-kinase (PI3K)-AKT (AKT serine/threonine kinase) that induced canonical transcription factors ZEB1 (zinc finger E-box binding homeobox 1) and SNAI1 (Snail family transcriptional repressor 1) and an EMT transcriptome. NFE2L2 deficiency disrupted RMNS that paradoxically normalized morphology but decreased function and viability. Thus, RPE heterogeneity is defined by the interaction of two cytoprotective pathways that is triggered by mitophagy function. By neutralizing the consequences of impaired mitophagy, an antioxidant dendrimer tropic for the RPE and mitochondria, EMT (a recognized AMD alteration) was abrogated to offer potential therapy for early AMD, a stage without treatment.Abbreviations: ACTB: actin beta; AKT: AKT serine/threonine kinase; AMD: age-related macular degeneration; CCCP: cyanide m-chlorophenyl hydrazone; CDH1: cadherin 1; DAVID: Database for Annotation, Visualization and Integrated Discovery; DHE: dihydroethidium; D-NAC: N-acetyl-l-cysteine conjugated to a poly(amido amine) dendrimer; ECAR: extracellular acidification rate; EMT: epithelial-mesenchymal transition; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GSEA: Gene Set Enrichment Analysis; HSPD1: heat shock protein family D (Hsp60) member 1; IVT: intravitreal; KD: knockdown; LMNA, lamin A/C; MAP1LC3B: microtubule associated protein 1 light chain 3 beta; MMP: mitochondrial membrane potential; NAC: N-acetyl-l-cysteine; NQO1: NAD(P)H quinone dehydrogenase 1; NFE2L2: NFE2 like bZIP transcription factor 2; O2-: superoxide anion; OCR: oxygen consumption rate; PI3K: phosphoinositide 3-kinase; PINK1: PTEN induced kinase 1; RMNS: retrograde mitochondrial-nuclear signaling; ROS: reactive oxygen species; RPE: retinal pigment epithelium; SNAI1: snail family transcriptional repressor 1; TJP1: tight junction protein 1; TPP-D-NAC: triphenyl phosphinium and N-acetyl-l-cysteine conjugated to a poly(amido amine) dendrimer; TIMM23: translocase of inner mitochondrial membrane 23; TOMM20: translocase of outer mitochondrial membrane 20; Trig: trigonelline; TXNRD1: thioredoxin reductase 1; VIM: vimentin; WT: wild-type; ZEB1: zinc finger E-box binding homeobox 1.
Keywords: NFE2L2; PINK1; age-related macular degeneration; dendrimer; epithelial mesenchymal transition; heterogeneity; mitophagy; retinal pigment epithelium; retrograde mitochondrial-nuclear signaling.
Conflict of interest statement
JTH: Bayer Pharmaceuticals (received grant funding and royalties on unrelated project); Clover Pharmaceuticals (receives grant funding and for being on the Scientific Advisory Board on unrelated project), Seeing Medicines, Inc. (receives payment for being on Scientific Advisory Board on unrelated project)
KR: R.K. and his wife S. Kannan are co-founders and own shares of Ashvattha Therapeutics Inc. They have licensed more than 80 patents into Ashvattha and serve in the Board of Directors. As part of the licensing of inventions, they may receive royalties from sales of products. The conflict of interest is managed by Johns Hopkins University.
DS: receives grant funding from Astellas Pharma Inc., F. Hoffmann-La Roche Ltd., Generian Pharmaceuticals Inc, and UPMC Enterprises.
Figures



References
-
- Wong WL, Su X, Li X, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2(2):e106–16. - PubMed
-
- Handa JT. New molecular histopathologic insights into the pathogenesis of age-related macular degeneration. Int Ophthalmol Clin. 2007;47(1):15–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
