Atomic-Scale View of Protein-PEG Interactions that Redirect the Thermal Unfolding Pathway of PEGylated Human Galectin-3
- PMID: 35922375
- PMCID: PMC9529833
- DOI: 10.1002/anie.202203784
Atomic-Scale View of Protein-PEG Interactions that Redirect the Thermal Unfolding Pathway of PEGylated Human Galectin-3
Abstract
PEGylation is a promising approach to address the central challenge of applying biologics, i.e., lack of protein stability in the demanding environment of the human body. Wider application is hindered by lack of atomic level understanding of protein-PEG interactions, preventing design of conjugates with predicted properties. We deployed an integrative structural and biophysical approach to address this critical challenge with the PEGylated carbohydrate recognition domain of human galectin-3 (Gal3C), a lectin essential for cell adhesion and potential biologic. PEGylation dramatically increased Gal3C thermal stability, forming a stable intermediate and redirecting its unfolding pathway. Structural details revealed by NMR pointed to a potential role of PEG localization facilitated by charged residues. Replacing these residues subtly altered the protein-PEG interface and thermal unfolding behavior, providing insight into rationally designing conjugates while preserving PEGylation benefits.
Keywords: Biologics; NMR Spectroscopy; PEGylated Proteins; Protein Engineering; Protein-Polymer Conjugate.
© 2022 Wiley-VCH GmbH.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures
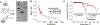

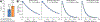
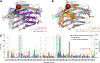

Similar articles
-
Conserved Protein-Polymer Interactions across Structurally Diverse Polymers Underlie Alterations to Protein Thermal Unfolding.ACS Cent Sci. 2023 Mar 14;9(4):685-695. doi: 10.1021/acscentsci.2c01522. eCollection 2023 Apr 26. ACS Cent Sci. 2023. PMID: 37122463 Free PMC article.
-
Analysis of monoPEGylated human galectin-2 by small-angle X-ray and neutron scattering: concentration dependence of PEG conformation in the conjugate.Biomacromolecules. 2010 Dec 13;11(12):3504-10. doi: 10.1021/bm100999a. Epub 2010 Nov 4. Biomacromolecules. 2010. PMID: 21049922
-
Chemical and Enzymatic Site Specific PEGylation of hGH: The Stability and in vivo Activity of PEG-N-Terminal-hGH and PEG-Gln141-hGH Conjugates.Macromol Biosci. 2016 Jan;16(1):50-6. doi: 10.1002/mabi.201500282. Epub 2015 Sep 9. Macromol Biosci. 2016. PMID: 26350165
-
The impact of PEGylation on biological therapies.BioDrugs. 2008;22(5):315-29. doi: 10.2165/00063030-200822050-00004. BioDrugs. 2008. PMID: 18778113 Review.
-
Purification of pegylated proteins.Methods Biochem Anal. 2011;54:339-62. doi: 10.1002/9780470939932.ch14. Methods Biochem Anal. 2011. PMID: 21954785 Review.
Cited by
-
Context-dependent effect of polyethylene glycol on the structure and dynamics of hirudin.Biophys J. 2025 Jan 7;124(1):192-204. doi: 10.1016/j.bpj.2024.11.3311. Epub 2024 Nov 26. Biophys J. 2025. PMID: 39600093
-
Conserved Protein-Polymer Interactions across Structurally Diverse Polymers Underlie Alterations to Protein Thermal Unfolding.ACS Cent Sci. 2023 Mar 14;9(4):685-695. doi: 10.1021/acscentsci.2c01522. eCollection 2023 Apr 26. ACS Cent Sci. 2023. PMID: 37122463 Free PMC article.
References
-
- Anselmo AC, Gokarn Y, Mitragotri S, Nat. Rev. Drug Discov 2019, 18, 19–40. - PubMed
-
- Harris JM, Chess RB, Nat. Rev. Drug Discov 2003, 2, 214–221. - PubMed
-
- Krall N, Cruz F. P. d., Boutureira O, Bernardes GJL, Nat. Chem 2016, 8, 103–113. - PubMed
-
- Shaunak S, Godwin A, Choi J-W, Balan S, Pedone E, Vijayarangam D, Heidelberger S, Teo I, Zloh M, Brocchini S, Nat. Chem. Biol 2006, 2, 312–313. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

