Methods to capture proteomic and metabolomic signatures from cerebrospinal fluid and serum of healthy individuals
- PMID: 35922450
- PMCID: PMC9349260
- DOI: 10.1038/s41598-022-16598-1
Methods to capture proteomic and metabolomic signatures from cerebrospinal fluid and serum of healthy individuals
Abstract
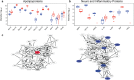
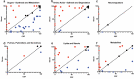
Discovery of reliable signatures for the empirical diagnosis of neurological diseases-both infectious and non-infectious-remains unrealized. One of the primary challenges encountered in such studies is the lack of a comprehensive database representative of a signature background that exists in healthy individuals, and against which an aberrant event can be assessed. For neurological insults and injuries, it is important to understand the normal profile in the neuronal (cerebrospinal fluid) and systemic fluids (e.g., blood). Here, we present the first comparative multi-omic human database of signatures derived from a population of 30 individuals (15 males, 15 females, 23-74 years) of serum and cerebrospinal fluid. In addition to empirical signatures, we also assigned common pathways between serum and CSF. Together, our findings provide a cohort against which aberrant signature profiles in individuals with neurological injuries/disease can be assessed-providing a pathway for comprehensive diagnostics and therapeutics discovery.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

