Chromatographic separation of glycated peptide isomers derived from glucose and fructose
- PMID: 35922676
- PMCID: PMC9436859
- DOI: 10.1007/s00216-022-04243-9
Chromatographic separation of glycated peptide isomers derived from glucose and fructose
Abstract
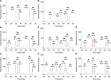
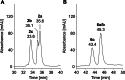
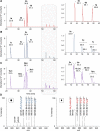
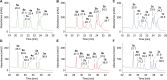
Amino groups in proteins can react with aldehyde groups in aldoses or keto groups in ketoses, e.g., D-glucose and D-fructose, yielding Schiff bases that rearrange to more stable Amadori and Heyns products, respectively. Analytical strategies to identify and quantify each glycation product in the presence of the corresponding isomer are challenged by similar physicochemical properties, impeding chromatographic separations, and by identical masses including very similar fragmentation patterns in tandem mass spectrometry. Thus, we studied the separation of seven peptide families, each consisting of unmodified, glucated, and fructated 15mer to 22mer peptides using reversed-phase (RP) and hydrophilic interaction chromatography (HILIC). In RP-HPLC using acidic acetonitrile gradients, unglycated peptides eluted ~ 0.1 to 0.8 min after the corresponding glycated peptides with four of seven peptides being baseline separated. Isomeric glucated and fructated peptides typically coeluted, although two late-eluting peptides were partially separated. Neutral eluents (pH 7.2) improved the chromatographic resolution (Rs), especially in the presence of phosphate, providing good and often even baseline separations for six of the seven isomeric glycated peptide pairs with fructated peptides eluting earlier (Rs = 0.7 to 1.5). Some glucated and unmodified peptides coeluted, but they can be distinguished by mass spectrometry. HILIC separated glycated and unmodified peptides well, whereas glucated and fructated peptides typically coeluted. In conclusion, HILIC efficiently separated unmodified and the corresponding glycated peptides, while isomeric Amadori and Heyns peptides were best separated by RP-HPLC using phosphate buffered eluents.
Keywords: Amadori and Heyns peptides; Fructation; Glucation; Hydrophilic interaction chromatography (HILIC); Reversed-phase high-performance liquid chromatography (RP-HPLC).
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
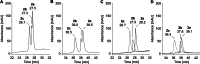
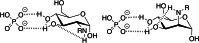
Similar articles
-
Differentiation and Quantitation of Coeluting Isomeric Amadori and Heyns Peptides Using Sugar-Specific Fragment Ion Ratios.Anal Chem. 2022 Jun 7;94(22):7909-7917. doi: 10.1021/acs.analchem.2c00681. Epub 2022 May 24. Anal Chem. 2022. PMID: 35609340
-
Solid-phase synthesis of D-fructose-derived Heyns peptides utilizing Nα-Fmoc-Lysin[Nε-(2-deoxy-D-glucos-2-yl),Nε-Boc]-OH as building block.Amino Acids. 2021 Jun;53(6):881-891. doi: 10.1007/s00726-021-02989-7. Epub 2021 May 2. Amino Acids. 2021. PMID: 33934222 Free PMC article.
-
Isomer selectivity of one- and two-dimensional approaches of mixed-mode and hydrophilic interaction liquid chromatography coupled to tandem mass spectrometry for sugar phosphates of glycolysis and pentose phosphate pathways.J Chromatogr A. 2023 Jan 11;1688:463727. doi: 10.1016/j.chroma.2022.463727. Epub 2022 Dec 18. J Chromatogr A. 2023. PMID: 36566570
-
Separation of Amadori peptides from their unmodified analogs by ion-pairing RP-HPLC with heptafluorobutyric acid as ion-pair reagent.Anal Bioanal Chem. 2008 Nov;392(6):1209-14. doi: 10.1007/s00216-008-2377-1. Epub 2008 Sep 24. Anal Bioanal Chem. 2008. PMID: 18813915
-
Peptide separation by Hydrophilic-Interaction Chromatography: a review.J Biochem Biophys Methods. 2004 Sep 30;60(3):265-80. doi: 10.1016/j.jbbm.2004.01.006. J Biochem Biophys Methods. 2004. PMID: 15345295 Review.
Cited by
-
Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2021-2022.Mass Spectrom Rev. 2025 May-Jun;44(3):213-453. doi: 10.1002/mas.21873. Epub 2024 Jun 24. Mass Spectrom Rev. 2025. PMID: 38925550 Free PMC article. Review.
-
The NMR signature of maltose-based glycation in full-length proteins.J Biomol NMR. 2024 Mar;78(1):61-72. doi: 10.1007/s10858-023-00432-5. Epub 2023 Dec 20. J Biomol NMR. 2024. PMID: 38114873 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

