Circular RNA expression profiles and CircSnd1-miR-135b/c-foxl2 axis analysis in gonadal differentiation of protogynous hermaphroditic ricefield eel Monopterus albus
- PMID: 35922747
- PMCID: PMC9347082
- DOI: 10.1186/s12864-022-08783-3
Circular RNA expression profiles and CircSnd1-miR-135b/c-foxl2 axis analysis in gonadal differentiation of protogynous hermaphroditic ricefield eel Monopterus albus
Abstract
Background: The expression and biological functions of circular RNAs (circRNAs) in reproductive organs have been extensively reported. However, it is still unclear whether circRNAs are involved in sex change. To this end, RNA sequencing (RNA-seq) was performed in gonads at 5 sexual stages (ovary, early intersexual stage gonad, middle intersexual stage gonad, late intersexual stage gonad, and testis) of ricefield eel, and the expression profiles and potential functions of circRNAs were studied.
Results: Seven hundred twenty-one circRNAs were identified, and the expression levels of 10 circRNAs were verified by quantitative real-time PCR (qRT-PCR) and found to be in accordance with the RNA-seq data, suggesting that the RNA-seq data were reliable. Then, the sequence length, category, sequence composition and the relationship between the parent genes of the circRNAs were explored. A total of 147 circRNAs were differentially expressed in the sex change process, and GO and KEGG analyses revealed that some differentially expressed (such as novel_circ_0000659, novel_circ_0004005 and novel_circ_0005865) circRNAs were closely involved in sex change. Furthermore, expression pattern analysis demonstrated that both circSnd1 and foxl2 were downregulated in the process of sex change, which was contrary to mal-miR-135b. Finally, dual-luciferase reporter assay and RNA immunoprecipitation showed that circSnd1 and foxl2 can combine with mal-miR-135b and mal-miR-135c. These data revealed that circSnd1 regulates foxl2 expression in the sex change of ricefield eel by acting as a sponge of mal-miR-135b/c.
Conclusion: Our results are the first to demonstrate that circRNAs have potential effects on sex change in ricefield eel; and circSnd1 could regulate foxl2 expression in the sex change of ricefield eel by acting as a sponge of mal-miR-135b/c. These data will be useful for enhancing our understanding of sequential hermaphroditism and sex change in ricefield eel or other teleosts.
Keywords: Expression pattern; Foxl2; Gonadal differentiation; Monopterus albus; Reproductive; circRNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

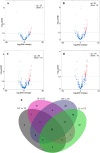


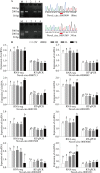
 : represents back-splicing sites. B Verification of circRNA expression patterns. The circRNA expression level of RNA-seq was calculated as Log50(TPM), and the RT–qPCR data were computed as the mean ± S.E.M. TPM: transcripts per million
: represents back-splicing sites. B Verification of circRNA expression patterns. The circRNA expression level of RNA-seq was calculated as Log50(TPM), and the RT–qPCR data were computed as the mean ± S.E.M. TPM: transcripts per million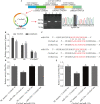


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

