Eukaryotic Microbial RNA Viruses-Acute or Persistent? Insights into Their Function in the Aquatic Ecosystem
- PMID: 35922920
- PMCID: PMC9763035
- DOI: 10.1264/jsme2.ME22034
Eukaryotic Microbial RNA Viruses-Acute or Persistent? Insights into Their Function in the Aquatic Ecosystem
Abstract
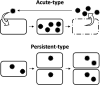
Isolated RNA viruses mainly parasitize eukaryotes. RNA viruses either expand horizontally by infecting hosts (acute type) or coexist with the host and are vertically inherited (persistent type). The significance of persistent-type RNA viruses in environmental viromes (the main hosts are expected to be microbes) was only recently reported because they had previously been overlooked in virology. In this review, we summarize the host-virus relationships of eukaryotic microbial RNA viruses. Picornavirales and Reoviridae are recognized as representative acute-type virus families, and most of the microbial viruses in Narnaviridae, Totiviridae, and Partitiviridae are categorized as representative persistent-type viruses. Acute-type viruses have only been found in aquatic environments, while persistent-type viruses are present in various environments, including aquatic environments. Moreover, persistent-type viruses are potentially widely spread in the RNA viral sequence space. This emerging evidence provides novel insights into RNA viral diversity, host-virus relationships, and their history of co-evolution.
Keywords: RNA virus; aquatic; eukaryote.
Figures


Similar articles
-
RNA Viruses in Aquatic Unicellular Eukaryotes.Viruses. 2021 Feb 25;13(3):362. doi: 10.3390/v13030362. Viruses. 2021. PMID: 33668994 Free PMC article. Review.
-
RNA Viruses in Aquatic Ecosystems through the Lens of Ecological Genomics and Transcriptomics.Viruses. 2022 Mar 28;14(4):702. doi: 10.3390/v14040702. Viruses. 2022. PMID: 35458432 Free PMC article. Review.
-
Viromes in marine ecosystems reveal remarkable invertebrate RNA virus diversity.Sci China Life Sci. 2022 Feb;65(2):426-437. doi: 10.1007/s11427-020-1936-2. Epub 2021 Jun 17. Sci China Life Sci. 2022. PMID: 34156600
-
Diverse RNA Viruses Associated with Diatom, Eustigmatophyte, Dinoflagellate, and Rhodophyte Microalgae Cultures.J Virol. 2022 Oct 26;96(20):e0078322. doi: 10.1128/jvi.00783-22. Epub 2022 Oct 3. J Virol. 2022. PMID: 36190242 Free PMC article.
-
RNA viromes from terrestrial sites across China expand environmental viral diversity.Nat Microbiol. 2022 Aug;7(8):1312-1323. doi: 10.1038/s41564-022-01180-2. Epub 2022 Jul 28. Nat Microbiol. 2022. PMID: 35902778
Cited by
-
The direct and indirect drivers shaping RNA viral communities in grassland soils.mSystems. 2024 Aug 20;9(8):e0009924. doi: 10.1128/msystems.00099-24. Epub 2024 Jul 9. mSystems. 2024. PMID: 38980057 Free PMC article.
-
Greetings from virologists to mycologists: A review outlining viruses that live in fungi.Mycoscience. 2024 Jan 31;65(1):1-11. doi: 10.47371/mycosci.2023.11.004. eCollection 2024. Mycoscience. 2024. PMID: 39239117 Free PMC article.
-
The First Identification of a Narnavirus in Bigyra, a Marine Protist.Microbes Environ. 2023;38(1):ME22077. doi: 10.1264/jsme2.ME22077. Microbes Environ. 2023. PMID: 36858534 Free PMC article.
-
Identification of shared viral sequences in peat moss metagenomes reveals elements of a possible Sphagnum core virome.Environ Microbiome. 2025 Jun 5;20(1):62. doi: 10.1186/s40793-025-00719-0. Environ Microbiome. 2025. PMID: 40474246 Free PMC article.
-
Efficient elimination of RNA mycoviruses in aspergillus species using RdRp-inhibitors ribavirin and 2'-C-methylribonucleoside derivatives.Front Microbiol. 2022 Oct 6;13:1024933. doi: 10.3389/fmicb.2022.1024933. eCollection 2022. Front Microbiol. 2022. PMID: 36274709 Free PMC article.
References
-
- Aihara, M., Urayama, S., Le, M.T., Katoh, Y., Higashiura, T., Fukuhara, T., et al. (2018) Infection by Magnaporthe oryzae chrysovirus 1 strain A triggers reduced virulence and pathogenic race conversion of its host fungus, Magnaporthe oryzae. J Gen Plant Pathol 84: 92–103.
-
- Banks, G.T., Buck, K.W., Chain, E.B., Darbyshire, J.E., and Himmelweit, F. (1969) Virus-like particles in penicillin producing strains of Penicillium chrysogenum. Nature 222: 89–90. - PubMed
-
- Bao, X., and Roossinck, M.J. (2013) Multiplexed interactions: viruses of endophytic fungi. In Advances in Virus Research. Amsterdam: Elsevier, pp. 37–58. - PubMed
-
- Beijerinck, M.W. (1898) Concerning a contagium vivum fluidum as cause of the spot disease of tobacco leaves. Phytopathol Classics 7: 33–52.

