LINC01234 Accelerates the Progression of Breast Cancer via the miR-525-5p/Cold Shock Domain-Containing E1 Axis
- PMID: 35923244
- PMCID: PMC9343190
- DOI: 10.1155/2022/6899777
LINC01234 Accelerates the Progression of Breast Cancer via the miR-525-5p/Cold Shock Domain-Containing E1 Axis
Abstract
Backgrounds: Long noncoding RNAs (lncRNAs) are strongly associated with the development of breast cancer (BC). As yet, the function of LINC01234 in BC remains unknown.
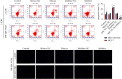
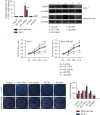
Methods: Using biological information, the potential lncRNA, miRNA, and target gene were predicted. LINC01234 and miR-525-5p expression in BC tissues was detected using quantitative real-time reverse transcription polymerase chain reaction. Fluorescence in situ hybridization was used to determine the distribution of LINC01234. Cell proliferation was analyzed using CCK-8 assay, colony formation, terminal deoxynucleotidyl transferase dUTP nick end labeling staining, and apoptosis evaluated using flow cytometry. Western blotting was used to evaluate protein expression. Dual-luciferase® reporter, RNA pull-down, and RNA immunoprecipitation assays were performed to analyze the binding relationships among LINC01234, miR-525-5p, and cold shock domain-containing E1 (CSDE1).
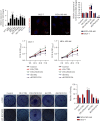
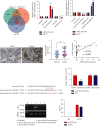
Results: We screened out LINC01234, found to be significantly increased in BC tissues, associated with a poor prognosis, and positively correlated with tumor size of BC. Knockdown of LINC01234 suppressed BC cell growth and facilitated apoptosis. Dual-luciferase reporter®, RNA pull-down, and RNA immunoprecipitation assays confirmed that LINC01234 and CSDE1 directly interacted with miR-525-5p. Upregulation of miR-525-5p and suppression of CSDE1 inhibited BC cell growth and induced cell apoptosis.
Conclusion: Upregulation of LINC01234 contributes to the development of BC through the miR-525-5p/CSDE1 axis. LINC01234 may be one of the potential diagnostic and treatment targets for BC.
Copyright © 2022 Jia Yu et al.
Conflict of interest statement
The authors have no relevant financial or nonfinancial interests to disclose.
Figures





Similar articles
-
Long Noncoding RNA NEAT1 Promotes the Progression of Breast Cancer by Regulating miR-138-5p/ZFX Axis.Cancer Biother Radiopharm. 2022 Oct;37(8):636-649. doi: 10.1089/cbr.2019.3515. Epub 2020 Aug 21. Cancer Biother Radiopharm. 2022. PMID: 32833504
-
Long noncoding RNA LINC01234 silencing exerts an anti-oncogenic effect in esophageal cancer cells through microRNA-193a-5p-mediated CCNE1 downregulation.Cell Oncol (Dordr). 2020 Jun;43(3):377-394. doi: 10.1007/s13402-019-00493-5. Epub 2020 Mar 4. Cell Oncol (Dordr). 2020. PMID: 32130660
-
LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer.Cancer Med. 2018 Mar;7(3):842-855. doi: 10.1002/cam4.1353. Epub 2018 Feb 23. Cancer Med. 2018. Retraction in: Cancer Med. 2023 Dec;12(24):22437. doi: 10.1002/cam4.6773. PMID: 29473328 Free PMC article. Retracted.
-
The lncRNA HOXA11-AS acts as a tumor promoter in breast cancer through regulation of the miR-125a-5p/TMPRSS4 axis.J Gene Med. 2022 May;24(5):e3413. doi: 10.1002/jgm.3413. Epub 2022 Feb 27. J Gene Med. 2022. PMID: 35106863
-
Oncogenic roles of LINC01234 in various forms of human cancer.Biomed Pharmacother. 2022 Oct;154:113570. doi: 10.1016/j.biopha.2022.113570. Epub 2022 Aug 26. Biomed Pharmacother. 2022. PMID: 36030582 Review.
Cited by
-
CSDE1: a versatile regulator of gene expression in cancer.NAR Cancer. 2024 Apr 10;6(2):zcae014. doi: 10.1093/narcan/zcae014. eCollection 2024 Jun. NAR Cancer. 2024. PMID: 38600987 Free PMC article.
-
Ensemble-based classification using microRNA expression identifies a breast cancer patient subgroup with an ultralow long-term risk of metastases.Cancer Med. 2024 May;13(9):e7089. doi: 10.1002/cam4.7089. Cancer Med. 2024. PMID: 38676390 Free PMC article.
-
Multi-transcriptomics predicts clinical outcome in systemically untreated breast cancer patients with extensive follow-up.Breast Cancer Res. 2025 Jul 15;27(1):133. doi: 10.1186/s13058-025-02061-2. Breast Cancer Res. 2025. PMID: 40665339 Free PMC article.
-
Diagnostic and prognostic role of microRNA-525 in different cancers: a systematic review and meta-analysis.Transl Cancer Res. 2024 Aug 31;13(8):4301-4314. doi: 10.21037/tcr-24-383. Epub 2024 Aug 27. Transl Cancer Res. 2024. PMID: 39262458 Free PMC article.
-
MiR-525-5p inhibits diffuse large B cell lymphoma progression via the Myd88/NF-κB signaling pathway.PeerJ. 2023 Nov 6;11:e16388. doi: 10.7717/peerj.16388. eCollection 2023. PeerJ. 2023. PMID: 37953776 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

