Performance of Multiple Pretrained BERT Models to Automate and Accelerate Data Annotation for Large Datasets
- PMID: 35923377
- PMCID: PMC9344209
- DOI: 10.1148/ryai.220007
Performance of Multiple Pretrained BERT Models to Automate and Accelerate Data Annotation for Large Datasets
Abstract
Purpose: To develop and evaluate domain-specific and pretrained bidirectional encoder representations from transformers (BERT) models in a transfer learning task on varying training dataset sizes to annotate a larger overall dataset.
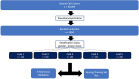
Materials and methods: The authors retrospectively reviewed 69 095 anonymized adult chest radiograph reports (reports dated April 2020-March 2021). From the overall cohort, 1004 reports were randomly selected and labeled for the presence or absence of each of the following devices: endotracheal tube (ETT), enterogastric tube (NGT, or Dobhoff tube), central venous catheter (CVC), and Swan-Ganz catheter (SGC). Pretrained transformer models (BERT, PubMedBERT, DistilBERT, RoBERTa, and DeBERTa) were trained, validated, and tested on 60%, 20%, and 20%, respectively, of these reports through fivefold cross-validation. Additional training involved varying dataset sizes with 5%, 10%, 15%, 20%, and 40% of the 1004 reports. The best-performing epochs were used to assess area under the receiver operating characteristic curve (AUC) and determine run time on the overall dataset.
Results: The highest average AUCs from fivefold cross-validation were 0.996 for ETT (RoBERTa), 0.994 for NGT (RoBERTa), 0.991 for CVC (PubMedBERT), and 0.98 for SGC (PubMedBERT). DeBERTa demonstrated the highest AUC for each support device trained on 5% of the training set. PubMedBERT showed a higher AUC with a decreasing training set size compared with BERT. Training and validation time was shortest for DistilBERT at 3 minutes 39 seconds on the annotated cohort.
Conclusion: Pretrained and domain-specific transformer models required small training datasets and short training times to create a highly accurate final model that expedites autonomous annotation of large datasets.Keywords: Informatics, Named Entity Recognition, Transfer Learning Supplemental material is available for this article. ©RSNA, 2022See also the commentary by Zech in this issue.
Keywords: Informatics; Named Entity Recognition; Transfer Learning.
© 2022 by the Radiological Society of North America, Inc.
Conflict of interest statement
Disclosures of conflicts of interest: A.S.T. No relevant relationships. Y.S.N. No relevant relationships. Y.X. No relevant relationships. J.R.F. No relevant relationships. T.G.B. Consulting fees from Change Healthcare. J.C.R. No relevant relationships.
Figures




References
-
- Hripcsak G , Austin JH , Alderson PO , Friedman C . Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports . Radiology 2002. ; 224 ( 1 ): 157 – 163 . - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

